For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNHEMPTGPIPVRRPLVPRMVRVLAVPIIVFWAFLAVTTNTFVPQVEKVAEELAGPMIPHYAPSQRAMLH IGEKFQESTSTSLTMVVLESQDRVLGDEDHRYYDELMQRFKDDSEHVQYVMDLWGKSITSAGAQSIDGKS TYVLLRLAGDIGQMQANESVRAVRAIVDKQTPPAGLRVYVSGPAPLAGDTLDIANSSLNNITIITIILII VMLLLVYRSFSTLLLPLAGVLIEMLVTKGVIATLGHLGYIELSSFAVNIVVSLTLGAGTDYGIFLLGRYH EARSTGQSREEAYYTAYRGVAPVVIGSGLTIAGACYCLSFARLNYFHTMGPAVAIAMVLTIAAALTLVPA VLTVGSLFGLFDPKAKAKAQLYRRIGASVVRWPVPIFVASSIIVMVGAVFVPTYQVSYDDRAYQPAGAPA NQGFAAADRHFSKSKLFTEMLMVESDHDMRNSADFISLDRVAKSLVRLPGVSMVQSITRPLGRPLEHATL PYLFTTQGSGNGQQLPFTKRQNGNTAQQAEIMGNTIVVLRDMIDLTQKLTDAMHATVVTMEDMQQVTEAM DQYVSNLDDFMRPLRNYFYWEPHCFDIPACWAFRSLFDMLDSVDKLAADIKDAVAQLEIVDKLLPQMVSQ LKVMANDMEGIRALIINTYGTSDLQSTLTTQTFDDMINVGLDFDKSRSDDFFYIPRQGFENEDVKTGMQL LMSPDGMAARFMITHEGNAMGPEGVEHVEQFPEAVGLALKETSLAGARVYVGGAGSNDKDIKEYGASDLL IVAIAAFVLIFLIMMFLTRSLMAPFVIIGTVAFSYSGAFGLSVLIWQHLIGLPLHWLVLPLTFIILVAVG SDYNLLLVARLKEETGAGLNTGLIRALGSTGAVVTSAGLVFAFTMLAMLASDLRTIGQVGSTICIGLLLD TLIVRSFIVPSLVRLFGTWFWWPTRVFARPLRRQNVGAHR*
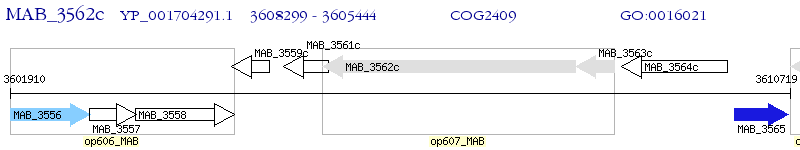
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3562c | - | - | 100% (951) | MmpL family protein |
| M. abscessus ATCC 19977 | MAB_4768c | - | 0.0 | 80.24% (931) | putative membrane protein, MmpL |
| M. abscessus ATCC 19977 | MAB_2037 | - | 0.0 | 55.47% (932) | putative membrane protein MmpL |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0458c | mmpL4 | 0.0 | 48.66% (933) | transmembrane transport protein MmpL4 |
| M. gilvum PYR-GCK | Mflv_3435 | - | 0.0 | 47.28% (939) | transport protein |
| M. tuberculosis H37Rv | Rv0450c | mmpL4 | 0.0 | 48.77% (933) | transmembrane transport protein MmpL4 |
| M. leprae Br4923 | MLBr_02378 | - | 0.0 | 48.33% (927) | hypothetical protein MLBr_02378 |
| M. marinum M | MMAR_3744 | mmpL4_1 | 0.0 | 48.78% (945) | transmembrane transport protein MmpL4_1 |
| M. avium 104 | MAV_3863 | - | 0.0 | 47.25% (927) | MmpL4 protein |
| M. smegmatis MC2 155 | MSMEG_0185 | - | 0.0 | 73.12% (930) | MmpL6 protein |
| M. thermoresistible (build 8) | TH_3056 | - | 0.0 | 44.46% (929) | PUTATIVE Mmp14a protein |
| M. ulcerans Agy99 | MUL_3688 | mmpL4_1 | 0.0 | 48.57% (945) | transmembrane transport protein MmpL4_1 |
| M. vanbaalenii PYR-1 | Mvan_3189 | - | 0.0 | 46.96% (939) | transport protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0458c|M.bovis_AF2122/97 ------------------MSTKFANDSNTNARPEKPFIARMIHAFAVPII
Rv0450c|M.tuberculosis_H37Rv ------------------MSTKFANDSNTNARPEKPFIARMIHAFAVPII
MLBr_02378|M.leprae_Br4923 ------------------MTVKCANDLDTHTKP--PFVARMIHRFAVPII
Mflv_3435|M.gilvum_PYR-GCK --------------MSAPVSDARTDEFPAAHGPHRSFIPRMIRLFAVPII
Mvan_3189|M.vanbaalenii_PYR-1 --------------MSAPVSDARTDEFPAARQPHRPFIPRMIRLFAVPII
TH_3056|M.thermoresistible__bu ------------------MSVLDR----VGVEPRRPRVASFIRRYSVLII
MAV_3863|M.avium_104 ---------------------------MTTDQATRSVVAQTIRRLSVPIL
MMAR_3744|M.marinum_M MSESNSGASISTEDTGPIEIGAATGGAPNRERGHRPVIPHTIRILAVPII
MUL_3688|M.ulcerans_Agy99 MSESNSGASISTEDTGPIEIGAATGGAPNRERGHRPVIPHTIRILAVPII
MAB_3562c|M.abscessus_ATCC_199 --------------------MSNHEMPTGPIPVRRPLVPRMVRVLAVPII
MSMEG_0185|M.smegmatis_MC2_155 --------------------MP-GQVRMSAHRAGRPFFARTVRVLAVPII
. .. :: :* *:
Mb0458c|M.bovis_AF2122/97 LGWLAVCVVVTVFVPSLEAVGQERSVSLSPKDAPSFEAMGRIGMVFKEGD
Rv0450c|M.tuberculosis_H37Rv LGWLAVCVVVTVFVPSLEAVGQERSVSLSPKDAPSFEAMGRIGMVFKEGD
MLBr_02378|M.leprae_Br4923 LVWLAIAVTVSVFIPSLEDVGQERSVSLSPKDAPSYIAMQKMGQVFNEGN
Mflv_3435|M.gilvum_PYR-GCK LGWIALIVVVNVTVPQLETVGEAQAVSMSPNDAPSMISMKKVGELFEEGD
Mvan_3189|M.vanbaalenii_PYR-1 LGWIALIVILNVTVPQLEAVGEARAVSMSPNEAPSLISMKKVGELFREGD
TH_3056|M.thermoresistible__bu LGWVALTVLATVAVPPLEAVEREKSLSLIPYDAPAFAAMHRMGELFDEPD
MAV_3863|M.avium_104 LVWVAVAAISNIAVPNLEDVAKAHNVSLNTPAAPSFQAMQHIGKVFHEYD
MMAR_3744|M.marinum_M LGWVLLTVLVNVVVPSLEVVSEQHSAPMTPMDAPSMKAMMRLGHNFREFD
MUL_3688|M.ulcerans_Agy99 LGWVLLTVLVNVVVPSLEVVSEQHSAPMTPMDAPSMKAMMRLGHNFREFD
MAB_3562c|M.abscessus_ATCC_199 VFWAFLAVTTNTFVPQVEKVAEELAGPMIPHYAPSQRAMLHIGEKFQEST
MSMEG_0185|M.smegmatis_MC2_155 VFWGLLAVTTNTFIPQVEHVADELAGPMIPNYAPSQVALLHIGEKFQESD
: * : . . :* :* * .: . **: :: ::* * *
Mb0458c|M.bovis_AF2122/97 SDSFAMVIIEG-NQPLGDAAHKYYDGLVAQLRADKKHVQSVQDLWGDPLT
Rv0450c|M.tuberculosis_H37Rv SDSFAMVIIEG-NQPLGDAAHKYYDGLVAQLRADKKHVQSVQDLWGDPLT
MLBr_02378|M.leprae_Br4923 SDSVIMIVLEG-DKPLGDDAHRFYDGLIRKLRADK-HVQSVQDFWGDPLT
Mflv_3435|M.gilvum_PYR-GCK SDSSVMIVFEG-DQPLGDEAHAWYDELVDRLEADTAHVQSVQDFWSDPLT
Mvan_3189|M.vanbaalenii_PYR-1 SDSSVMIVFEG-DQPLGDEAHAWYDELVERLRADTKHVQSVQDFWSDPLT
TH_3056|M.thermoresistible__bu SESIAIIVLEG-QEPLGLEAHEYYDQLISQLREHPQYVQHIRDFWGDPLT
MAV_3863|M.avium_104 SDSAAMIVLEG-DKPLGDDAHRFYDALVGKLEHDTKHVEHVQNFWGDPLT
MMAR_3744|M.marinum_M SNSTVMIVLEG-QHKLGDDAHQYYDSLIRQLRQDPEHIQHIQDFWGDRLT
MUL_3688|M.ulcerans_Agy99 SNSTVMIVLEG-QHKLGDDAHQYYDSLIRQLRQDPEHIQHIQDFWGDRLT
MAB_3562c|M.abscessus_ATCC_199 STSLTMVVLESQDRVLGDEDHRYYDELMQRFKDDSEHVQYVMDLWGKSIT
MSMEG_0185|M.smegmatis_MC2_155 STNLTMLVFEA-DRPLDDGDHRYYDDLVRRLQADTEHVQYVMDLWGKPIT
* . ::::*. :. *. * :** *: ::. . ::: : ::*.. :*
Mb0458c|M.bovis_AF2122/97 AAGVQSNDGKAAYVQLSLAGNQGTPLANESVEAVRSIVESTPAPPGIKAY
Rv0450c|M.tuberculosis_H37Rv AAGVQSNDGKAAYVQLSLAGNQGTPLANESVEAVRSIVESTPAPPGIKAY
MLBr_02378|M.leprae_Br4923 APGAQSNDGKAAYVQLSLAGNQGELLAQESIDAVRKIVVQTPAPPGITAW
Mflv_3435|M.gilvum_PYR-GCK ASGSQSNDGKAAYVQVKLSGNQGEALANESVKAVQQTVSSLEPPPGVRAF
Mvan_3189|M.vanbaalenii_PYR-1 ASGSQSNDGKAAYVQVKLAGNQGESLANESVQAAQEIVRSLEPPPGVRAF
TH_3056|M.thermoresistible__bu AGAAQSDDGKAAFVQVNLVGRFGQAKVNEAVRAVQEIVERTPAPEGVETF
MAV_3863|M.avium_104 AAGSQSKDGKAAYVQVYLAGNQGDALANESVDAIRNIVEHTPPPPGVKAY
MMAR_3744|M.marinum_M AAGAQSADGKSAYVMVNLAGNQGTTLANESVEAVRDVINRTHEPAGVKAY
MUL_3688|M.ulcerans_Agy99 AAGAQSADGKSAYVMVNLAGNQGTTLANESVEAVRDVINRTHEPAGVKAY
MAB_3562c|M.abscessus_ATCC_199 SAGAQSIDGKSTYVLLRLAGDIGQMQANESVRAVRAIVDKQTPPAGLRVY
MSMEG_0185|M.smegmatis_MC2_155 AAGAQSVDGKASFVLLRLAGNIGQLEANESVDAVRDIIADDAPPQGLKVY
: . ** ***:::* : * * * .:*:: * : : * *: .:
Mb0458c|M.bovis_AF2122/97 VTGPSALAADMHHSGDRSMARITMVTVAVIFIMLLLVYRSIITVVLLLIT
Rv0450c|M.tuberculosis_H37Rv VTGPSALAADMHHSGDRSMARITMVTVAVIFIMLLLVYRSIITVVLLLIT
MLBr_02378|M.leprae_Br4923 VTGASALIADMHHSGDKSMIRITATSVIVILVTLLLVYRSFITVILLLFT
Mflv_3435|M.gilvum_PYR-GCK VTGPAALAADQHIAGDRSMRLIEAATFTVIIVMLLLVYRSIVTVLITLVM
Mvan_3189|M.vanbaalenii_PYR-1 VTGPAALAADQHIASDRSVRVIELVTFAVIIVMLLLVYRSIVTVLLTLVM
TH_3056|M.thermoresistible__bu VTGPAALGADAGDSGTATVILITLVTVAVIFLMLLFFFRSFLTVIVLLLT
MAV_3863|M.avium_104 VTGAAPLVTDQFEVGSKGIFKVTVITVLVILAMLLWVYRS-VTAVFVLVT
MMAR_3744|M.marinum_M VTGPAALSDDMHLIGNASLAKITMFTLGAIAVMLLLVYRSIVTTVIQLFM
MUL_3688|M.ulcerans_Agy99 VTGPAALSDDMHLIGNASLATITMFTLGAIAVMLLLVYRSIVTTVIQLFM
MAB_3562c|M.abscessus_ATCC_199 VSGPAPLAGDTLDIANSSLNNITIITIILIIVMLLLVYRSFSTLLLPLAG
MSMEG_0185|M.smegmatis_MC2_155 VSGAAPLASDTLSTANSSLNNITIFTIILIVVMLLLVYRSAACLLVPLPA
*:*.:.* * . : : :. * ** .:** :. *
Mb0458c|M.bovis_AF2122/97 VGVELTAARGVVAVLGHSGAIGLTTFAVSLLTSLAIAAGTDYGIFIIGRY
Rv0450c|M.tuberculosis_H37Rv VGVELTAARGVVAVLGHSGAIGLTTFAVSLLTSLAIAAGTDYGIFIIGRY
MLBr_02378|M.leprae_Br4923 VGIESAVARGVVALLGHTGLIGLSTFAVNLLTSLAIAAGTDYGIFITGRY
Mflv_3435|M.gilvum_PYR-GCK VVMSLATARGVVAFLGHHEIIGLSTFATNLLVTLAIAAATDYAIFLIGRY
Mvan_3189|M.vanbaalenii_PYR-1 VVLSLATARGVVAFLGWHEIIGLSLFATNLLVTLAIAAATDYAIFLIGRY
TH_3056|M.thermoresistible__bu VAVELQVARGLVAYLGLQGWVGLTTYVVNLLVSVGIAAGTDYGIFFFGRY
MAV_3863|M.avium_104 VIVEMAAARGIVAFLGSVGLIGLSTYATNLLTLLVIAAGTDYAIFFVGRY
MMAR_3744|M.marinum_M TGIALASSRGVVAVLGYHNVFGLTTFAANILTMLAIAAGTDYGIFLVGRY
MUL_3688|M.ulcerans_Agy99 TGIALASSRGVVAVLGYHNVFGLTTFAANILTMLAIAAGTDYGIFLVGRY
MAB_3562c|M.abscessus_ATCC_199 VLIEMLVTKGVIATLGHLGYIELSSFAVNIVVSLTLGAGTDYGIFLLGRY
MSMEG_0185|M.smegmatis_MC2_155 VLIEMLVAKGVIATLGHFGLIPLSSFAVNVVVALTLGAGTDYGIFLLGRY
. : ::*::* ** . *: :...::. : :.*.***.**: ***
Mb0458c|M.bovis_AF2122/97 QEARQAGEDKEAAYYTMYRGTAHVILGSGLTITGATFCLSFARMPYFQTL
Rv0450c|M.tuberculosis_H37Rv QEARQAGEDKEAAYYTMYRGTAHVILGSGLTIAGATFCLSFARMPYFQTL
MLBr_02378|M.leprae_Br4923 QEARQANENKEAAFYTMYRGTFHVILGSGLTISGATFCLSFARMPYFQTL
Mflv_3435|M.gilvum_PYR-GCK QEARTAGEDKETAYYTMFHGTAHVVLGSGLTIAGATFCLSFTRLPYFQTM
Mvan_3189|M.vanbaalenii_PYR-1 QEARTNGEDKESAYYTMFHGTAHVVLGSGLTIAGATYCLSFTRLPYFQTL
TH_3056|M.thermoresistible__bu QEAREAGEDRDSAYFTTYRSVAKVVLASGLTIAGAIACLSFTRLPHFQPL
MAV_3863|M.avium_104 HEARHEGQDRETAYYTMYRGTTHVVLGSGLTVAGAVLCLRFTRLNYFQSL
MMAR_3744|M.marinum_M QEARQAGEDREDAYYTTFRGVAPVVLGSGLTIAGATYCLSFARLPWFNTM
MUL_3688|M.ulcerans_Agy99 QEARQAGEDRQDAYYTTFRGVAPVVLGSGLTIAGATYCLSFARLPWFNTM
MAB_3562c|M.abscessus_ATCC_199 HEARSTGQSREEAYYTAYRGVAPVVIGSGLTIAGACYCLSFARLNYFHTM
MSMEG_0185|M.smegmatis_MC2_155 HEARQSGESREDAYFTAYRSVAPIIIGSGLTIAGACFCLSLARLDYFHTM
:*** .:.:: *::* ::.. :::.****::** ** ::*: *:.:
Mb0458c|M.bovis_AF2122/97 GIPCAVGMLVAVAVALTLGPAVLHVGSRFG-LFDPKRLLK-VRGWRRVGT
Rv0450c|M.tuberculosis_H37Rv GIPCAVGMLVAVAVALTLGPAVLHVGSRFG-LFDPKRLLK-VRGWRRVGT
MLBr_02378|M.leprae_Br4923 GVPCAVGMLIAVAVALTLGPAVLTVGSRFG-LFEPKRLIK-VRGWRRIGT
Mflv_3435|M.gilvum_PYR-GCK GVPLAIGMFVVVMAGVVLMVACISVATRFGKLLEPKRAMR-IRGWRKIGA
Mvan_3189|M.vanbaalenii_PYR-1 GVPLAIGMFVVVMAGVILMVAMISVATRFGKLLEPKRAMR-IRGWRKIGA
TH_3056|M.thermoresistible__bu GIPGAVGLLVAVAVALTLVPATIALGSRFG-LFEPKRRVM-THRWRRIGT
MAV_3863|M.avium_104 GIPAAIGIGVALAAALSLTPAVITVGSLFG-LFDPKRQMA-TRGWRRIGT
MMAR_3744|M.marinum_M GAPVAIGMLVVVLAGLTLGPAVVFVGSRFH-LFESKRAANRGRLWRRVGT
MUL_3688|M.ulcerans_Agy99 GAPVAIGMLVVVLAGLTLGPAVVFVGSRFH-PFESKRAANRGRLWRRVGT
MAB_3562c|M.abscessus_ATCC_199 GPAVAIAMVLTIAAALTLVPAVLTVGSLFG-LFDPKAKAK-AQLYRRIGA
MSMEG_0185|M.smegmatis_MC2_155 GPAVAISMLFTIAAALTLAPALLTLGSLLA-LFDPKRPTR-GHLYRRIGT
* . *:.: ..: ..: * * : :.: : ::.* : :*::*:
Mb0458c|M.bovis_AF2122/97 VVVRWPLPVLVATCAIALVGLLALPGYKTSYNDRDYLPDFIPANQGYAAA
Rv0450c|M.tuberculosis_H37Rv VVVRWPLPVLVATCAIALVGLLALPGYKTSYNDRDYLPDFIPANQGYAAA
MLBr_02378|M.leprae_Br4923 VVVRWPLPILITTCAIAMVGLLALPGYRTNYKDRAYLPASIPANQGFAAA
Mflv_3435|M.gilvum_PYR-GCK AIVRWPGPILVATMAITLVGLLALPGYKTNYNDRDYLPADLPANQGYAVA
Mvan_3189|M.vanbaalenii_PYR-1 AVVRWPGPILVATLAITLVGLLALPGYKTNYNDRTYLPADLPANEGYAVA
TH_3056|M.thermoresistible__bu AVVRWPVPILTAAVALSLVGLLTLPGYNPSYSDQEFIPDDIPASEGYKAA
MAV_3863|M.avium_104 AIVRWPGPILVVATGVALVGLLALPGYRTNYDARPYMPASAPANTGYTAA
MMAR_3744|M.marinum_M AVVRWPAPILALSGAIVLVGMVALPGFKTSYNDRYYLPASAPSNLGEAAA
MUL_3688|M.ulcerans_Agy99 AVVRWPAPILALSGAIVLVGMVALPGFKTSYNDRYYLPASAPSNLGEAAA
MAB_3562c|M.abscessus_ATCC_199 SVVRWPVPIFVASSIIVMVGAVFVPTYQVSYDDRAYQPAGAPANQGFAAA
MSMEG_0185|M.smegmatis_MC2_155 SVVRWPKPIFVASAAVVMVGAVFVPSYRVNYDDRAYQPVDAAANLGFQAA
:**** *:: : : :** : :* :. .*. : : * .:. * .*
Mb0458c|M.bovis_AF2122/97 DRHFSQARMK-PEILMIESDHDMRNPADFLVLDKLAKGIFRVPGISRVQA
Rv0450c|M.tuberculosis_H37Rv DRHFSQARMK-PEILMIESDHDMRNPADFLVLDKLAKGIFRVPGISRVQA
MLBr_02378|M.leprae_Br4923 DRHFPQARMK-PEILMIESDHDMRNPADFLILDKLARGIFRVPGISRVQA
Mflv_3435|M.gilvum_PYR-GCK DEHFDQARMN-PELLMIESDHDLRNSADFLVIDKIAKALFRVEGIARVQA
Mvan_3189|M.vanbaalenii_PYR-1 DRHFDQARMN-PELLMIESDHDLRNSADFLVIDKIAKAIFKVEGIARVQA
TH_3056|M.thermoresistible__bu ARHFPASAMSVPDVVLIEADRDMRNPADLLVLNKVAKKIFEVPGVANVQS
MAV_3863|M.avium_104 ERHFSQARLN-PELLMIETDHDMRNPADMINLERVAKAIAHLPGIAQVQS
MMAR_3744|M.marinum_M DRHFSHARMN-PDMLMVEAEHDMRNPADMLVLDKVAKNVIRTVGIAMIQD
MUL_3688|M.ulcerans_Agy99 DRHFSHARMN-PDMLMVEAEHDMRNPADMLVLDKVAKNVIRTVGIAMIQD
MAB_3562c|M.abscessus_ATCC_199 DRHFSKSKLF-TEMLMVESDHDMRNSADFISLDRVAKSLVRLPGVSMVQS
MSMEG_0185|M.smegmatis_MC2_155 DRHFNQSKLF-SEMLMIETDHDMRNSADFISLDRVAKALIRLPGVAMVQS
.** : : .:::::*:::*:**.**:: ::::*: : . *:: :*
Mb0458c|M.bovis_AF2122/97 ITRPEGTTMDHTSIPFQISMQNAGQLQTIKYQRDRANDMLKQADEMATTI
Rv0450c|M.tuberculosis_H37Rv ITRPEGTTMDHTSIPFQISMQNAGQLQTIKYQRDRANDMLKQADEMATTI
MLBr_02378|M.leprae_Br4923 ITRPDGTAMDHTSIPFQISMQNAGQVQTMKYQKDRMNDLLRQAENMAETI
Mflv_3435|M.gilvum_PYR-GCK ITRPDGKPIKHTSIPFQMSMQGTTQRLNEKYMQDRMADMLVQADEMANTI
Mvan_3189|M.vanbaalenii_PYR-1 ITRPDGKPIKHTSIPFQMSMQGTTQRLNEKYMQDRMADMLVQADEMANTI
TH_3056|M.thermoresistible__bu VTRPEGRQIEHTTIPFMLSMSQASQRLALPFQRARMDDMLKQADELTETI
MAV_3863|M.avium_104 MTRPLGTPIEHTSLAFQISAGSIGSIENLQYQKERAEDLLKQADNLKDTI
MMAR_3744|M.marinum_M ITRPLGIPIQHSSIPFQNSMQSQTMMQNMAFLKERMADMTKMADEMQVLI
MUL_3688|M.ulcerans_Agy99 ITRPLGIPIQHSSIPFQNSMQSQTMMQNMAFLKERMADMTKMADEMQVLI
MAB_3562c|M.abscessus_ATCC_199 ITRPLGRPLEHATLPYLFTTQGSGNGQQLPFTKRQNGNTAQQAEIMGNTI
MSMEG_0185|M.smegmatis_MC2_155 ITRPLGRPLEHATIPYLFTTQGSTNGQQLPFSEQANENTDRQAEIQARSV
:*** * :.*:::.: : : . : *: :
Mb0458c|M.bovis_AF2122/97 AVLTRMHSLMAEMASTTHRMVGDTEEMKEITEELRDHVADFDDFWRPIRS
Rv0450c|M.tuberculosis_H37Rv AVLTRMHSLMAEMASTTHRMVGDTEEMKEITEELRDHVADFDDFWRPIRS
MLBr_02378|M.leprae_Br4923 ASMRRMHQLMALLTENTHHILNDTVEMQKTTSKLRDEIANFDDFWRPIRS
Mflv_3435|M.gilvum_PYR-GCK KTMEKMSGLTAQMADITHSMVTKMEGMVVDIEDLRDSIANFDDFFRPIRN
Mvan_3189|M.vanbaalenii_PYR-1 ATMEKMSNLTAQMADITHSMVSKMENMLTDIEDLRDSIANFDDFFRPIRN
TH_3056|M.thermoresistible__bu ALMRRMQSLMEKMVATTHSMVGTTHELLDTTRDLRDHIADFDDFFRPIRN
MAV_3863|M.avium_104 GILNQQYALQKQLAASTHDETQSFHDTIAIINDLRDKIANFDDFFRPVRS
MMAR_3744|M.marinum_M DTTERMYAVMRDLDTTTTDMARVTAETSTITDELRDHLAEFDDFWRPMRS
MUL_3688|M.ulcerans_Agy99 DTTERMYAVMRELDTTTTDMARVTAETSTITDELRDHLAEFDDFWRPMRS
MAB_3562c|M.abscessus_ATCC_199 VVLRDMIDLTQKLTDAMHATVVTMEDMQQVTEAMDQYVSNLDDFMRPLRN
MSMEG_0185|M.smegmatis_MC2_155 EVLQHVITITQQMSAELHSTVLTFENLKEVTDEVNEGISNLDDFLRPLKN
: : : : ::::*** **::.
Mb0458c|M.bovis_AF2122/97 YFYWEKHCYGIPICWSFRSIFDALDGIDKLSEQIGVLLGDLREMDRLMPQ
Rv0450c|M.tuberculosis_H37Rv YFYWEKHCYGIPICWSFRSIFDALDGIDKLSEQIGVLLGDLREMDRLMPQ
MLBr_02378|M.leprae_Br4923 YFYWERHCFNIPICWSFRSIFDALDGVDQIDERLNSIVGDIKNMDLLMPQ
Mflv_3435|M.gilvum_PYR-GCK YFYWEPHCYNIPVCWALRSVFDTLDGINIMTDDFKAIIPDMKRLDTLMPQ
Mvan_3189|M.vanbaalenii_PYR-1 YFYWEPHCYNIPVCWALRSVFDTLDGINVMTDDFREILPDMKRLDSLMPQ
TH_3056|M.thermoresistible__bu YFHWEPHCYNIPICWSIRSIFDALDGINEMTEKMEELVEDMDRLDALMPQ
MAV_3863|M.avium_104 YFYWEKHCFDIPACFAFRSVFDALDGIDELSAQFEKLTASLDKLDAGQQK
MMAR_3744|M.marinum_M YFYWEKHCFDVPICWSFRSLFDSLDGIDKLSEKFSELTEDIQHTAGLTSQ
MUL_3688|M.ulcerans_Agy99 YFYWEKHCFDVPICWSFRSLFDSLDGIDKLSEKFSELTEDIQRTAGLTSQ
MAB_3562c|M.abscessus_ATCC_199 YFYWEPHCFDIPACWAFRSLFDMLDSVDKLAADIKDAVAQLEIVDKLLPQ
MSMEG_0185|M.smegmatis_MC2_155 YFYWEPHCFDIPVCFAMRSLFDSLDGIDSLDEQIGNAVVSFEAIDRLLPQ
**:** **:.:* *:::**:** **.:: : : .: :
Mb0458c|M.bovis_AF2122/97 MVAQIPPQIEAMENMRTMILTMHSTMTGIFDQMLEMSDNATAMGKAFDAA
Rv0450c|M.tuberculosis_H37Rv MVAQIPPQIEAMENMRTMILTMHSTMTGIFDQMLEMSDNATAMGKAFDAA
MLBr_02378|M.leprae_Br4923 MLEQFPPMIESMESMRTIMLTMHSTMSGIFDQMNELSDNANTMGKAFDTA
Mflv_3435|M.gilvum_PYR-GCK MVALMPEMIETMKTMRTMMLTMYQSQKGQQDQMAAMSEDADAMGEAFDDS
Mvan_3189|M.vanbaalenii_PYR-1 MVALMPEMIETMKTMRTMMLTMYQSQKGQQDQMAAMSEDADAMGEAFDDS
TH_3056|M.thermoresistible__bu MLVQFPQMIETMESSRTMILTMHATMSGIFDQMESSSENATAMGKAFDAS
MAV_3863|M.avium_104 LVTLLPPQIADQEKNLALTLSNYATNLGINAQTRANTDTATALGQAYDAA
MMAR_3744|M.marinum_M MLTLIPPMIATMKTTKALTLTMQSTFSAMLDQMDELSNTAIVMGQSFDAS
MUL_3688|M.ulcerans_Agy99 MLTLIPPMIATMKTTKALTLTMQSTFSAMLDQMDELSNTAIVMGQSFDAS
MAB_3562c|M.abscessus_ATCC_199 MVSQLKVMANDMEGIRALIINTYGTSDLQSTLTTQTFDDMINVGLDFDKS
MSMEG_0185|M.smegmatis_MC2_155 MITQLERMIADSRALLGVITNNYGPAHLQSTQTDQTYYDQINVGNDFDAA
:: : . : . . :* :* :
Mb0458c|M.bovis_AF2122/97 KNDDSFYLPPEVFKNKDFQRAMKSFLSSDGHAARFIILHRGDPQSPEGIK
Rv0450c|M.tuberculosis_H37Rv KNDDSFYLPPEVFKNKDFQRAMKSFLSSDGHAARFIILHRGDPQSPEGIK
MLBr_02378|M.leprae_Br4923 KNDDSFYLPPEVFKNTDFKRAMKSFLSSDGHAARFIILHRGDPASVAGIA
Mflv_3435|M.gilvum_PYR-GCK MNDDSFYLPPEIFENADFQRGLEQFLSPDGHAVRFIISHEGDPLSPEGVA
Mvan_3189|M.vanbaalenii_PYR-1 MNDDSFYLPPEIFENKDFQRGLEQFLSPDGHAVRFIISHEGDPLSPEGVA
TH_3056|M.thermoresistible__bu HNDDSFYLPPEALENKDFQRVMEIFLSPDGKAARMLVTQRGDPATPEGMA
MAV_3863|M.avium_104 KNDDSFYLPPEAFTNPEFKRGLKLFLSPDGKAARMIISHEGDPATPEGIS
MMAR_3744|M.marinum_M KNDDFFYLPPEAFDNPDFQTGLRMFLSPDGKSARFFITHQGDPMTPEGIS
MUL_3688|M.ulcerans_Agy99 KNDDFFYLPPEAFDNPDFQTGLRMFLSPDGKSARFFITHQGDPMTPEGIS
MAB_3562c|M.abscessus_ATCC_199 RSDDFFYIPRQGFENEDVKTGMQLLMSPDGMAARFMITHEGNAMGPEGVE
MSMEG_0185|M.smegmatis_MC2_155 RSDDFFYIPHEAFDNDDVKTGMKLLMSPDGKAARFIITHEGNAMAPEGIE
.** **:* : : * :.: :. ::*.** :.*::: :.*:. *:
Mb0458c|M.bovis_AF2122/97 SIDAIRTAAEESLKGTPLEDAKIYLAGTAAVFHDISEGAQWDLLIAAISS
Rv0450c|M.tuberculosis_H37Rv SIDAIRTAAEESLKGTPLEDAKIYLAGTAAVFHDISEGAQWDLLIAAISS
MLBr_02378|M.leprae_Br4923 SINAIRTAAEEALKGTPLEDTKIYLAGTAAVFKDIDEGANWDLVIAGISS
Mflv_3435|M.gilvum_PYR-GCK KIDKIKTAAKEAVKGTPLEGSKIYLGGTAAVFKDMQDGNNYDLLIAGIAA
Mvan_3189|M.vanbaalenii_PYR-1 KIDKIKTAAKEAIKGTPLEGSKIYLGGTAATFKDMQDGNNYDLLIAGISA
TH_3056|M.thermoresistible__bu LVEPIYIAAEEALKGTPLESSRIYLAGSAAGVKDLVEGAKYDLMIAAISA
MAV_3863|M.avium_104 HIEPIRNAAKQAVKNTPLGDSNIYLAGTAATYKDIAEGAKYDLLIAGIAA
MMAR_3744|M.marinum_M RVDAERTAAQEGLKQSSLSDAKVYLGGTAATFKDMADGEKYDLMIAVVSA
MUL_3688|M.ulcerans_Agy99 RVDAERTAAQEGLKQSSLSDAKVYLGGTAATFKDMADGEKYDLMIAVVSA
MAB_3562c|M.abscessus_ATCC_199 HVEQFPEAVGLALKETSLAGARVYVGGAGSNDKDIKEYGASDLLIVAIAA
MSMEG_0185|M.smegmatis_MC2_155 HVEQFPDAIKMALKETSLAGAKIYIGGAASNNLDIKTYAASDLLIVAIAA
:: * .:* :.* .:.:*:.*:.: *: **:*. :::
Mb0458c|M.bovis_AF2122/97 LCLIFIIMLIITRAFIAAAVIVGTVALSLGASFGLSVLLWQHILAIHLHW
Rv0450c|M.tuberculosis_H37Rv LCLIFIIMLIITRAFIAAAVIVGTVALSLGASFGLSVLLWQHILAIHLHW
MLBr_02378|M.leprae_Br4923 LCLIFIIMLIITRAFVAAAVIVGTVALSLGASFGLSVLLWQHILGIELHY
Mflv_3435|M.gilvum_PYR-GCK LGLIFIIMLILTRAVVAAAVIVGTVVLSLAASFGLSVLFWQHILGQPLHW
Mvan_3189|M.vanbaalenii_PYR-1 LGLIFIIMLILTRAIVAAAVIVGTVVLSLAASFGLSVLFWQHILGTELHW
TH_3056|M.thermoresistible__bu VCLIFMIMVFMTRSVIAALVIVGTVVLSLGASFGMSVLLWQHLLGIQLHW
MAV_3863|M.avium_104 LSLILLIMVLITRSLVAAIVIVGTVALSLGASFGLSVLVWQDILGIKLYW
MMAR_3744|M.marinum_M LTLIFLIMLLLTRSVVAALVIVGTAASSIAASFGLSVLIWQDLFGIKIHW
MUL_3688|M.ulcerans_Agy99 LTLIFLIMLLLTRSVVAALVIVGTAASSIAASFGLSVLIWQDLFGIKIHW
MAB_3562c|M.abscessus_ATCC_199 FVLIFLIMMFLTRSLMAPFVIIGTVAFSYSGAFGLSVLIWQHLIGLPLHW
MSMEG_0185|M.smegmatis_MC2_155 FVLIFLIMLYLTRSLVAALVIPGTVAFSFAGAFGLSVMMWQHLIGLPLHW
. **::**: :**:.:*. ** **.. * ..:**:**:.**.::. :::
Mb0458c|M.bovis_AF2122/97 LVLAMSVIVLLAVGSDYNLLLVSRFKQEIGAGLKTGIIRSMGGTGKVVTN
Rv0450c|M.tuberculosis_H37Rv LVLAMSVIVLLAVGSDYNLLLVSRFKQEIGAGLKTGIIRSMGGTGKVVTN
MLBr_02378|M.leprae_Br4923 LVLAMSVIVLLAVGSDYNLLLVSRFKQEIQAGLKTGIIRSMGGTGKVVTN
Mflv_3435|M.gilvum_PYR-GCK MVLAMAVIILLAVGADYNLLLVSRLKEEIHAGIGTGIIRAMGGSGSVVTA
Mvan_3189|M.vanbaalenii_PYR-1 MVLAMAVIVLLAVGADYNLLLVSRLKEEIHAGIGTGIIRAMGGSGSVVTA
TH_3056|M.thermoresistible__bu VVLAMSVIVLLAVGSDYNLMLVSRMKEEIGAGLNTGIIRAMGGTGRVVTM
MAV_3863|M.avium_104 ICLALSVIILLAVGSDYNLLLISRFREEIHAGLNTGIIRSMAGSGAVVTS
MMAR_3744|M.marinum_M IVAALSVIILLAVGSDYNLLLVSRFREEIHAGLKTGIIRSMAGTGGVVTA
MUL_3688|M.ulcerans_Agy99 IVAALSVIILLAVGSDYNLLLVSRFREEIHAGLKTGIIRSMAGTGGVVTA
MAB_3562c|M.abscessus_ATCC_199 LVLPLTFIILVAVGSDYNLLLVARLKEETGAGLNTGLIRALGSTGAVVTS
MSMEG_0185|M.smegmatis_MC2_155 LILPLSFIILVAVGSDYNLLLISRVKEETGAGLNTGLIRALGSTGAVVTS
: .::.*:*:***:****:*::*.::* **: **:**::..:* ***
Mb0458c|M.bovis_AF2122/97 AGLVFAVTMASMAVSDLRVIGQVGTTIGLGLLFDTLIVRSFMTPSIAALL
Rv0450c|M.tuberculosis_H37Rv AGLVFAVTMASMAVSDLRVIGQVGTTIGLGLLFDTLIVRSFMTPSIAALL
MLBr_02378|M.leprae_Br4923 AGLVFAFTMASMVVSDLRVIGQVGTTIGLGLLFDTLIVRSFMMPSIAALL
Mflv_3435|M.gilvum_PYR-GCK AGLVFALTMMSMSVSELTVIGQVGTTIGLGLLFDTLVIRAFMTPSIAALL
Mvan_3189|M.vanbaalenii_PYR-1 AGLVFALTMMSMAVSELTVIGQVGTTIGLGLLFDTLVIRAFMTPSIAALL
TH_3056|M.thermoresistible__bu AGLVFAATMASMIVSDLLMIGQVGTTIALGLLFDTMVVRAFMTPSVAAIL
MAV_3863|M.avium_104 AGLVFAFTMASFVSASLLVLGQIGTTIALGLLFDTLIVRSFMTPSVAALL
MMAR_3744|M.marinum_M AGLVFAFTMASMLGSDLRVLGQFGSTVCIGLLLDTLIVRTLLMPSIATLL
MUL_3688|M.ulcerans_Agy99 AGLVFAFTMASMLGSDLRVLGQFGSTVCIGLLLDTLIVRTLLMPSIATLL
MAB_3562c|M.abscessus_ATCC_199 AGLVFAFTMLAMLASDLRTIGQVGSTICIGLLLDTLIVRSFIVPSLVRLF
MSMEG_0185|M.smegmatis_MC2_155 AGLVFAFTMLSMLVSDLRTIGQLGTTVCIGLLLDTMIVRSLVVPCLLRLL
****** ** :: :.* :**.*:*: :***:**:::*::: *.: ::
Mb0458c|M.bovis_AF2122/97 GRWFWWPLRVRSRPARTPTVPSETQPAGRPLAMSSDRLG-----------
Rv0450c|M.tuberculosis_H37Rv GRWFWWPLRVRSRPARTPTVPSETQPAGRPLAMSSDRLG-----------
MLBr_02378|M.leprae_Br4923 GRWFWWPQQGRTRPLLTVAAPVGLPPD-RSLVYSD---------------
Mflv_3435|M.gilvum_PYR-GCK GPWFWWPQRVRTRPVPAPWPRSGGLQSDPSEGVRA---------------
Mvan_3189|M.vanbaalenii_PYR-1 GPWFWWPQRVRTRPVPAPWPRPGGLQSDPSEGVKV---------------
TH_3056|M.thermoresistible__bu GRWFWWPQRVRTRPASAMLRPVGPRPLVRSLLLRD---------------
MAV_3863|M.avium_104 GRWFWWPQRVRPRPASTMLRPYGPRPAVRQLLLWEDGDPAVAPETPSAAR
MMAR_3744|M.marinum_M GRWFWWPQVVHPRGDNARPRVAAPTH------------------------
MUL_3688|M.ulcerans_Agy99 GRWFWWPQVVHPRGDNARPRVAAPTH------------------------
MAB_3562c|M.abscessus_ATCC_199 GTWFWWPTRVFARPLRRQNVGAHR--------------------------
MSMEG_0185|M.smegmatis_MC2_155 GPWFWWPTFIRQRPLRQAVRQRERADVV----------------------
* ***** *
Mb0458c|M.bovis_AF2122/97 ----
Rv0450c|M.tuberculosis_H37Rv ----
MLBr_02378|M.leprae_Br4923 ----
Mflv_3435|M.gilvum_PYR-GCK ----
Mvan_3189|M.vanbaalenii_PYR-1 ----
TH_3056|M.thermoresistible__bu ----
MAV_3863|M.avium_104 HSRG
MMAR_3744|M.marinum_M ----
MUL_3688|M.ulcerans_Agy99 ----
MAB_3562c|M.abscessus_ATCC_199 ----
MSMEG_0185|M.smegmatis_MC2_155 ----