For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLEVIDKGACSESHPVPLLFVHGAWHAAWCWDEHFLDFFADKGFRALAVSLRGHGNSPSPKPLRTCSLAD YLDDVDSVVDTLPTTPVLIGHSMGGFIVQKYLEAHQAPAGVLIASAPQRGSFAFTMRLTRRHPWLTTKGL ITGNALLPVGTPELARESFFSSQTPESDVVRYAARLNGESQRVAIDTIWMLPRPKRVTTPLLILGAECDR SILSKEVRTTANAYRTEAEIFPDMGHDMMLEPGWAAVAERIHTWLEMRDLRPHRGEPDQQETTPTG
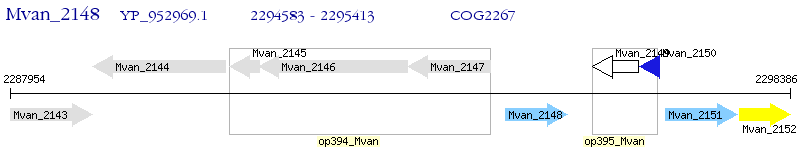
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2148 | - | - | 100% (276) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_3112 | - | 2e-91 | 62.69% (260) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_4177 | - | 1e-07 | 38.46% (78) | esterase EstC, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3502c | bpoA | 4e-06 | 31.65% (79) | peroxidase BpoA |
| M. gilvum PYR-GCK | Mflv_3379 | - | 4e-86 | 58.85% (260) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3473c | bpoA | 4e-06 | 31.65% (79) | peroxidase BpoA |
| M. leprae Br4923 | MLBr_02603 | - | 1e-09 | 26.19% (252) | hypothetical protein MLBr_02603 |
| M. abscessus ATCC 19977 | MAB_3517 | - | 4e-06 | 39.29% (84) | putative hydrolase, alpha/beta fold LipV |
| M. marinum M | MMAR_1669 | - | 1e-10 | 28.68% (272) | lysophospholipase |
| M. avium 104 | MAV_3771 | - | 6e-76 | 53.58% (265) | hydrolase, alpha/beta fold family protein, putative |
| M. smegmatis MC2 155 | MSMEG_2036 | - | 7e-10 | 25.67% (261) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_3727 | - | 2e-09 | 39.05% (105) | PUTATIVE Chloride peroxidase |
| M. ulcerans Agy99 | MUL_1905 | - | 3e-10 | 28.84% (267) | lysophospholipase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2148|M.vanbaalenii_PYR-1 ---------------MLEVIDKGACSES--HPVPLLFVHGAWHAAWCWDE
Mflv_3379|M.gilvum_PYR-GCK -----------MAPAPLEVIDKGESTDA--HPAPLLFVHGAWHAAWCWDD
MAV_3771|M.avium_104 ---------------MLEVIEKGSGSAE--HPTPLLFVHGGWHAAWCWEN
Mb3502c|M.bovis_AF2122/97 ----------------------------------MVFLHGGGQTRRSWGR
Rv3473c|M.tuberculosis_H37Rv ----------------------------------MVFLHGGGQTRRSWGR
MMAR_1669|M.marinum_M ------------------------------MRKHVVLIHGAWSRGRQLAG
MUL_1905|M.ulcerans_Agy99 ------------------------------MRKHVVLIHGAWSRGRQLAG
TH_3727|M.thermoresistible__bu ----------MATITTSDGVEIFYRDWG--SGQPIVFSHGWPLTADDWDP
MSMEG_2036|M.smegmatis_MC2_155 MTLPQLPPGRTFEVRAVDGVRLHAQTFGPEDGYPIVFAHGITCAIEVWAH
MAB_3517|M.abscessus_ATCC_1997 -----------------MTPTLNIHRYGPVEPARLLALHGLTGHGRRWAP
MLBr_02603|M.leprae_Br4923 --MTITRTERNFYGVGGVRIVYDVWMPDTRPRAVIILAHGFGEHARRYDH
:: **
Mvan_2148|M.vanbaalenii_PYR-1 HFLDFFADKGFRALAVSLRGHGNS--PSPKPLRTCSLADYLDDVDSVVDT
Mflv_3379|M.gilvum_PYR-GCK HFLDFFAGRGYRALALSFRGHGRS--ATDKPLRSLSISDYVDDVATVAAR
MAV_3771|M.avium_104 -FLDFFADAGYRAVALSLRGHGAS--PTSKPLHRVSIADYLDDVAAVAGE
Mb3502c|M.bovis_AF2122/97 -AAAAVAERGWQAVTIDLRGHGE---SDWSSEGDYRLVSFAGDIQEVLRN
Rv3473c|M.tuberculosis_H37Rv -AAAAVAERGWQAVTIDLRGHGE---SDWSSEGDYRLVSFAGDIQEVLRN
MMAR_1669|M.marinum_M -ARSAFEHHGYTVHTPTLRHHELPLPDGAARIATLSLRDYADDLVELVAS
MUL_1905|M.ulcerans_Agy99 -ARSAFEHHGYTVHTPTLRHHELPLPDGAARIATLSLRDYADDLVELVAS
TH_3727|M.thermoresistible__bu -QMLFFLNAGYRVIAHDRRGHGRS----SPVPDGHDMDHYADDLAALVEH
MSMEG_2036|M.smegmatis_MC2_155 QIADLAG--DYRVIAYDHRGHGRS--ETPRGRNRYSLNHLAADLDAVLDA
MAB_3517|M.abscessus_ATCC_1997 LFDEYLAD--VPVLAPDLLGHGRS-----PATAPWSLEAHADAVAAELDT
MLBr_02603|M.leprae_Br4923 -VAHYFAAAGLATYALDLRGHGRS---AGKRVLVRDLSEYNADFDILVGI
. : * : .
Mvan_2148|M.vanbaalenii_PYR-1 LPT-----TPVLIGHSMGGFIVQKYLEAHQ---APAGVLIASAPQ---RG
Mflv_3379|M.gilvum_PYR-GCK LPT-----PPIVVGHSMGGFVVQKYLEDRD---VPAGVLMASAPP---RG
MAV_3771|M.avium_104 LGG-----APILIGHSLGGFVIQRYLETHR---VPAAVLVGSVPP---QG
Mb3502c|M.bovis_AF2122/97 LPG-----QPALVGASLGGFAAMLLAGELSPGIASAVVLVDIVPNMDLAG
Rv3473c|M.tuberculosis_H37Rv LPG-----QPALVGASLGGFAAMLLAGELSPGIASAVVLVDIVPNMDLAG
MMAR_1669|M.marinum_M LDS-----APLLVGHSLGGLLVQLVAARTRHIGVVAACPSPVGP----VG
MUL_1905|M.ulcerans_Agy99 LNS-----APLLVGHSLGGLLVQLVAARTRHIGVVAACPSPVGP----VG
TH_3727|M.thermoresistible__bu LGLR----DAVHVGHSTGGGVVVRYLARHGEERTAKAVLIAAVPP-LMVR
MSMEG_2036|M.smegmatis_MC2_155 TLRPGE--RAVIAGHSMGGIAITSWAQRYPSRVQTCADGVALINTSTGDL
MAB_3517|M.abscessus_ATCC_1997 AETG----PIVVVGHSFGGAIALHLAARRPDLVKSLVLLDPAIGLDG-DW
MLBr_02603|M.leprae_Br4923 ATRDHPGLKRIVAGHSMGGAIVFAYGVERPDNYDLMVLSGPAVAAQDMVS
* * **
Mvan_2148|M.vanbaalenii_PYR-1 SFAFTMRLTRR--------HPWLTTKGLITGNALLPVGT---PELARESF
Mflv_3379|M.gilvum_PYR-GCK YLGSGVRWFRR--------HPWQFVRTSLSGESLAYVSP---IEAARERF
MAV_3771|M.avium_104 VLRLALRVWRR--------RPSMTMEAWNDPTLLKFLAT---PALAREYL
Mb3502c|M.bovis_AF2122/97 ASRIHAFMAERVESGFGSLDEVADVIANYNPHRPRPSDPDGLVANLRRRG
Rv3473c|M.tuberculosis_H37Rv ASRIHAFMAERVESGFGSLDEVADVIANYNPHRPRPSDPDGLVANLRRRG
MMAR_1669|M.marinum_M LNRTTAGLSMR-----HALRPRPWTKPVAPPSWPRFRTG---IAGAQSAG
MUL_1905|M.ulcerans_Agy99 LNRTTAGLSMR-----HALRPRPWTKPVAPPSWPRFRTG---IAGAQSAG
TH_3727|M.thermoresistible__bu TDANPGGLPKS------VFDDFQAQLAANRSEFYRALASGPFYGFNRDGV
MSMEG_2036|M.smegmatis_MC2_155 LRDVQLAQVPTRLSTTRIRTAGTILKTFGSTPVPKLADAANRRFVAHLAV
MAB_3517|M.abscessus_ATCC_1997 MRQIADQMVTY--------PDYTDRAEAKSDKVAGAWSDVPDHVIEAELD
MLBr_02603|M.leprae_Br4923 PLRAVVGKGLG--------------LVAPGLPVHQLEVD----AISRNRA
Mvan_2148|M.vanbaalenii_PYR-1 FSSQTPESDVVRYAARLNGESQRVAI-DTIWMLP------RPKRVTTPLL
Mflv_3379|M.gilvum_PYR-GCK FSPSTPEAIVAACAARLQEESARSGR-DGVTALP------RPKRVRAPML
MAV_3771|M.avium_104 FCAATPEAIVESCRQRAGAESVRAAMTDPMLRRV------RTRRVSTPIL
Mb3502c|M.bovis_AF2122/97 DRWYWHWDPQFIGGIAAFPPVEVTDVDRMNAAVA------TILRDEVPVL
Rv3473c|M.tuberculosis_H37Rv DRWYWHWDPQFIGGIAAFPPVEVTDVDRMNAAVA------TILRDEVPVL
MMAR_1669|M.marinum_M AAREMFEDLVCESGRVLFFELALPWLDRSKVARV------DYPAVTGPVL
MUL_1905|M.ulcerans_Agy99 AAREMFDDLVCESGRVLFFELALPWLDRSKVARV------DYPAVTGPVL
TH_3727|M.thermoresistible__bu EPSEALIANWWRQGMMGDAKAHYDGIVAFSQTDFT----EDLTKITVPVL
MSMEG_2036|M.smegmatis_MC2_155 GRDADPWVADFVYRLFATTPPTGRGAWGRVLVDNLGPKHIPLHNLTVPTL
MAB_3517|M.abscessus_ATCC_1997 EHLIQLSNGRCGWRMNLPAAVTAWSELARDIVLP--------VNVIRTTL
MLBr_02603|M.leprae_Br4923 VVAAYKDDPLVYHGKVPAGVGRVMLQVGETMTRR-------APVLTTPLL
. *
Mvan_2148|M.vanbaalenii_PYR-1 ILGAECD-RSILSKEVRTTANAYR--TEAEIFPDMGHDMMLEPGWA-AVA
Mflv_3379|M.gilvum_PYR-GCK VLGALDDGMAVTPAEVHATARAYR--TEATLFPGMGHNMMLEPGWE-AVA
MAV_3771|M.avium_104 VLGATHD-GFVSAADVRATARAYR--TDPEFF-DMGHNMMLEPGWV-AVA
Mb3502c|M.bovis_AF2122/97 LVRGQVSDIVRQESADQFLSRFPQ--VEFTDVRGAGHMVAGDRND--AFA
Rv3473c|M.tuberculosis_H37Rv LVRGQVSDIVRQESADQFLSRFPQ--VEFTDVRGAGHMVAGDRND--AFA
MMAR_1669|M.marinum_M VLGGEYDRIVGSTIARQTAGRYRN--GTFVEIPGSDHLVFSARVLP-ATM
MUL_1905|M.ulcerans_Agy99 VLGGEYDRIVGSAIARQTAGRYRN--GTFVEIPGSDHLVFSARVLP-ATM
TH_3727|M.thermoresistible__bu VMHGDDDQVVPYANSGPLTAELLRN-AKLKTYQGFPHGMMT-THAE-VVN
MSMEG_2036|M.smegmatis_MC2_155 VIGSAQDRLLPINASRRIADTVPNL-AAFVELRGGHCGILERPDEVNAQL
MAB_3517|M.abscessus_ATCC_1997 LLATRTSPPYVLPKLLDALQATLG---SQFELIDIDCDHMVSQSRPAETA
MLBr_02603|M.leprae_Br4923 VVHGSEDRLVLVDGSHRLVECVGSTDVELKVYPGLYHEVFNEPERD-QVL
:: . .
Mvan_2148|M.vanbaalenii_PYR-1 ERIHTWLEMRDLRPHRGEPDQQETTPTG
Mflv_3379|M.gilvum_PYR-GCK SHIDSWLISRGL----------------
MAV_3771|M.avium_104 ERIRDWLQAPTAARHR------------
Mb3502c|M.bovis_AF2122/97 GAVLDFLARHVGVR--------------
Rv3473c|M.tuberculosis_H37Rv GAVLDFLARHVGVR--------------
MMAR_1669|M.marinum_M ARISEWIAHNQLFATA------------
MUL_1905|M.ulcerans_Agy99 ARISEWIAHNHLFATA------------
TH_3727|M.thermoresistible__bu ADILEFLRS-------------------
MSMEG_2036|M.smegmatis_MC2_155 RKLAEAVISPQRLNS-------------
MAB_3517|M.abscessus_ATCC_1997 AAIRSHLE--------------------
MLBr_02603|M.leprae_Br4923 EDVVCWILKRL-----------------
: :