For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LEVIDKGSVSQSHPVPLLFVHGAWHAAWCWDEHFLDFFAGRGYRALAVSLRGHGGSPADKKLRDLSFEDF VADITTAADALPTRPVIIGHSMGGVLVQRYLETRDAPAGVLMASMPPQGSLKSGLRWIRSHPWHFAKLTA TGRSLPYVSTPELARERFFSPATPESVVRHYAARLQEESARMGLDGLVKLPHPSRVRTPLLVLGAADDGA VTQDEVRATADAYGTEAHIFPGMGHNMMLEAGWEDVADRIDVWLRSRRL*
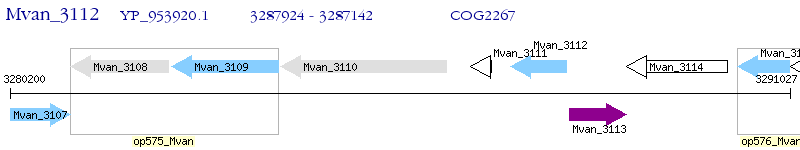
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3112 | - | - | 100% (260) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_2148 | - | 1e-91 | 62.69% (260) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_1567 | - | 3e-09 | 28.11% (217) | chloride peroxidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3502c | bpoA | 1e-05 | 31.07% (103) | peroxidase BpoA |
| M. gilvum PYR-GCK | Mflv_3379 | - | 1e-103 | 68.46% (260) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3473c | bpoA | 1e-05 | 31.07% (103) | peroxidase BpoA |
| M. leprae Br4923 | MLBr_02603 | - | 3e-10 | 24.90% (249) | hypothetical protein MLBr_02603 |
| M. abscessus ATCC 19977 | MAB_4551c | - | 6e-05 | 23.61% (233) | lysophospholipase |
| M. marinum M | MMAR_1669 | - | 5e-08 | 27.04% (270) | lysophospholipase |
| M. avium 104 | MAV_3771 | - | 1e-68 | 51.94% (258) | hydrolase, alpha/beta fold family protein, putative |
| M. smegmatis MC2 155 | MSMEG_0460 | - | 4e-07 | 41.25% (80) | alpha/beta hydrolase fold |
| M. thermoresistible (build 8) | TH_3727 | - | 3e-09 | 38.32% (107) | PUTATIVE Chloride peroxidase |
| M. ulcerans Agy99 | MUL_1905 | - | 2e-07 | 26.67% (270) | lysophospholipase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3112|M.vanbaalenii_PYR-1 --------------------------------------------MLEVID
Mflv_3379|M.gilvum_PYR-GCK ----------------------------------------MAPAPLEVID
MAV_3771|M.avium_104 --------------------------------------------MLEVIE
Mb3502c|M.bovis_AF2122/97 --------------------------------------------------
Rv3473c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1669|M.marinum_M --------------------------------------------------
MUL_1905|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02603|M.leprae_Br4923 -----------------------------MTITRTERNFYGVGGVRIVYD
MAB_4551c|M.abscessus_ATCC_199 ----------------------------------MQRTFDGADDVRIVYD
TH_3727|M.thermoresistible__bu ---------------------------------------MATITTSDGVE
MSMEG_0460|M.smegmatis_MC2_155 MRHADEVSPLFQRQRPTPSDPPEGTPEWFTAALEHRPQHSTIDFEGCRIH
Mvan_3112|M.vanbaalenii_PYR-1 KGSVSQSHPVPLLFVHGAWHAAWCWDEHFLDFFAGRGYRALAVSLRGHGG
Mflv_3379|M.gilvum_PYR-GCK KGESTDAHPAPLLFVHGAWHAAWCWDDHFLDFFAGRGYRALALSFRGHGR
MAV_3771|M.avium_104 KGSGSAEHPTPLLFVHGGWHAAWCWEN-FLDFFADAGYRAVALSLRGHGA
Mb3502c|M.bovis_AF2122/97 -----------MVFLHGGGQTRRSWGR-AAAAVAERGWQAVTIDLRGHGE
Rv3473c|M.tuberculosis_H37Rv -----------MVFLHGGGQTRRSWGR-AAAAVAERGWQAVTIDLRGHGE
MMAR_1669|M.marinum_M -------MRKHVVLIHGAWSRGRQLAG-ARSAFEHHGYTVHTPTLRHHEL
MUL_1905|M.ulcerans_Agy99 -------MRKHVVLIHGAWSRGRQLAG-ARSAFEHHGYTVHTPTLRHHEL
MLBr_02603|M.leprae_Br4923 VWMPDTRPRAVIILAHGFGEHARRYDH-VAHYFAAAGLATYALDLRGHGR
MAB_4551c|M.abscessus_ATCC_199 TWTPAGTPRAVVVLSHGFGEHARRYDH-VARRFNEAGYLVYALDHRGHGR
TH_3727|M.thermoresistible__bu IFYRDWGSGQPIVFSHGWPLTADDWDP-QMLFFLNAGYRVIAHDRRGHGR
MSMEG_0460|M.smegmatis_MC2_155 LRTWGDPENPPLVFVHGGAAHSGWWDH--IAPFFTRTHRVIAPDVSGHGD
::: ** . . : *
Mvan_3112|M.vanbaalenii_PYR-1 S--PADKKLRDLSFEDFVADITTAADALPTRP-----VIIGHSMGGVLVQ
Mflv_3379|M.gilvum_PYR-GCK S--ATDKPLRSLSISDYVDDVATVAARLPTPP-----IVVGHSMGGFVVQ
MAV_3771|M.avium_104 S--PTSKPLHRVSIADYLDDVAAVAGELGGAP-----ILIGHSLGGFVIQ
Mb3502c|M.bovis_AF2122/97 ---SDWSSEGDYRLVSFAGDIQEVLRNLPGQP-----ALVGASLGGFAAM
Rv3473c|M.tuberculosis_H37Rv ---SDWSSEGDYRLVSFAGDIQEVLRNLPGQP-----ALVGASLGGFAAM
MMAR_1669|M.marinum_M PLPDGAARIATLSLRDYADDLVELVASLDSAP-----LLVGHSLGGLLVQ
MUL_1905|M.ulcerans_Agy99 PLPDGAARIATLSLRDYADDLVELVASLNSAP-----LLVGHSLGGLLVQ
MLBr_02603|M.leprae_Br4923 ---SAGKRVLVRDLSEYNADFDILVGIATRDHPGLKRIVAGHSMGGAIVF
MAB_4551c|M.abscessus_ATCC_199 ---SGGKRVYLRDISEYTDDFGTLVDIAAREHPDLKRIVLGHSMGGGIVF
TH_3727|M.thermoresistible__bu ----SSPVPDGHDMDHYADDLAALVEHLGLRD----AVHVGHSTGGGVVV
MSMEG_0460|M.smegmatis_MC2_155 -----SGTRSHYSLTKWAREVLAVAAAEGPAAR---PTIVGHSMGGWVTA
: : :. * * **
Mvan_3112|M.vanbaalenii_PYR-1 RYLETRDAPAGVLMASMPPQGSLKSGLRWIRSHPWHFAK-----------
Mflv_3379|M.gilvum_PYR-GCK KYLEDRDVPAGVLMASAPPRGYLGSGVRWFRRHPWQFVR-----------
MAV_3771|M.avium_104 RYLETHRVPAAVLVGSVPPQGVLRLALRVWRRRPSMTME-----------
Mb3502c|M.bovis_AF2122/97 LLAGEL-SPGIASAVVLVDIVPNMDLAGASRIHAFMAERVESGFGSLDEV
Rv3473c|M.tuberculosis_H37Rv LLAGEL-SPGIASAVVLVDIVPNMDLAGASRIHAFMAERVESGFGSLDEV
MMAR_1669|M.marinum_M LVAART-RHIGVVAACPSPVGP----VGLNRTTAGLSMR-----HALRPR
MUL_1905|M.ulcerans_Agy99 LVAART-RHIGVVAACPSPVGP----VGLNRTTAGLSMR-----HALRPR
MLBr_02603|M.leprae_Br4923 AYGVER-PDNYDLMVLSGPAVAAQDMVSPLRAVVGKGLG-----------
MAB_4551c|M.abscessus_ATCC_199 AYGVDH-QDRYDLMVLSGPAIAAQVGLPYVLTLVAPVVG-----------
TH_3727|M.thermoresistible__bu RYLARHGEERTAKAVLIAAVPPLMVRTDANPGGLPKSVFD----------
MSMEG_0460|M.smegmatis_MC2_155 TAAMKYGADIDSILIIDSPLRERAPEEGRLRNRRSDHPG----------Y
Mvan_3112|M.vanbaalenii_PYR-1 -------LTATGRSLPYVSTPELARERFFSPATPESV--VRHYAARLQEE
Mflv_3379|M.gilvum_PYR-GCK -------TSLSGESLAYVSPIEAARERFFSPSTPEAI--VAACAARLQEE
MAV_3771|M.avium_104 -------AWNDPTLLKFLATPALAREYLFCAATPEAI--VESCRQRAGAE
Mb3502c|M.bovis_AF2122/97 ADVIANYNPHRPRPSDPDGLVANLRRRGDRWYWHWDP--QFIGGIAAFPP
Rv3473c|M.tuberculosis_H37Rv ADVIANYNPHRPRPSDPDGLVANLRRRGDRWYWHWDP--QFIGGIAAFPP
MMAR_1669|M.marinum_M PWTKPVAPPSWPRFRTG---IAGAQSAGAAREMFEDL--VCESGRVLFFE
MUL_1905|M.ulcerans_Agy99 PWTKPVAPPSWPRFRTG---IAGAQSAGAAREMFDDL--VCESGRVLFFE
MLBr_02603|M.leprae_Br4923 -----LVAPGLPVHQLEVD--AISRNRAVVAAYKDDP--LVYHG-KVPAG
MAB_4551c|M.abscessus_ATCC_199 -----RLAPGLPVQKLDVN--AISHDPAIIAAYHADP--LVHHG-RVPAG
TH_3727|M.thermoresistible__bu -DFQAQLAANRSEFYRALASGPFYGFNRDGVEPSEAL--IANWWRQGMMG
MSMEG_0460|M.smegmatis_MC2_155 PSREEILARFRAVPAQDVTLPFIDRHIASESVRQTDTGWVWKFDPAIFSG
Mvan_3112|M.vanbaalenii_PYR-1 SARMGL------DGLVKLPHPSRVRTPLLVLGAADDG-AVTQDEVRATAD
Mflv_3379|M.gilvum_PYR-GCK SARSGR------DGVTALPRPKRVRAPMLVLGALDDGMAVTPAEVHATAR
MAV_3771|M.avium_104 SVRAAMT-----DPMLRRVRTRRVSTPILVLGATHDG-FVSAADVRATAR
Mb3502c|M.bovis_AF2122/97 VEVTDVD-----RMNAAVATILRDEVPVLLVRGQVSD-IVRQESADQFLS
Rv3473c|M.tuberculosis_H37Rv VEVTDVD-----RMNAAVATILRDEVPVLLVRGQVSD-IVRQESADQFLS
MMAR_1669|M.marinum_M LALPWLD-----RSKVARVDYPAVTGPVLVLGGEYDR-IVGSTIARQTAG
MUL_1905|M.ulcerans_Agy99 LALPWLD-----RSKVARVDYPAVTGPVLVLGGEYDR-IVGSAIARQTAG
MLBr_02603|M.leprae_Br4923 VGRVMLQ-----VGETMTRRAPVLTTPLLVVHGSEDR-LVLVDGSHRLVE
MAB_4551c|M.abscessus_ATCC_199 IGRALLG-----VGKTMRQRAAGLKRPVLTMHGSDDR-LTAPEGSRWLSE
TH_3727|M.thermoresistible__bu DAKAHYDGIVAFSQTDFTEDLTKITVPVLVMHGDDDQVVPYANSGPLTAE
MSMEG_0460|M.smegmatis_MC2_155 SLQGQTP------AEQEILENTFAQIPCRVGYLRCENGLVPPDMAEQIRS
* .
Mvan_3112|M.vanbaalenii_PYR-1 AY---GTEAHIFPGMGHNMMLEAGWEDVADRIDVWLRSRRL----
Mflv_3379|M.gilvum_PYR-GCK AY---RTEATLFPGMGHNMMLEPGWEAVASHIDSWLISRGL----
MAV_3771|M.avium_104 AY---RTDPEFFD-MGHNMMLEPGWVAVAERIRDWLQAPTAARHR
Mb3502c|M.bovis_AF2122/97 RF--PQVEFTDVRGAGHMVAGDRND-AFAGAVLDFLARHVGVR--
Rv3473c|M.tuberculosis_H37Rv RF--PQVEFTDVRGAGHMVAGDRND-AFAGAVLDFLARHVGVR--
MMAR_1669|M.marinum_M RY--RNGTFVEIPGSDHLVFSARVLPATMARISEWIAHNQLFATA
MUL_1905|M.ulcerans_Agy99 RY--RNGTFVEIPGSDHLVFSARVLPATMARISEWIAHNHLFATA
MLBr_02603|M.leprae_Br4923 CVGSTDVELKVYPGLYHEVFNEPERDQVLEDVVCWILKRL-----
MAB_4551c|M.abscessus_ATCC_199 SA--PDATLKIWNGLYHEIFNEFEKELVLDEVVGWIDARL-----
TH_3727|M.thermoresistible__bu LLR--NAKLKTYQGFPHGMMT-THAEVVNADILEFLRS-------
MSMEG_0460|M.smegmatis_MC2_155 TLQLRGPFVELALAGHHPMLDQPLPLVATIRTLLEFWSIT-----
* : :