For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTPTLNIHRYGPVEPARLLALHGLTGHGRRWAPLFDEYLADVPVLAPDLLGHGRSPATAPWSLEAHADAV AAELDTAETGPIVVVGHSFGGAIALHLAARRPDLVKSLVLLDPAIGLDGDWMRQIADQMVTYPDYTDRAE AKSDKVAGAWSDVPDHVIEAELDEHLIQLSNGRCGWRMNLPAAVTAWSELARDIVLPVNVIRTTLLLATR TSPPYVLPKLLDALQATLGSQFELIDIDCDHMVSQSRPAETAAAIRSHLE
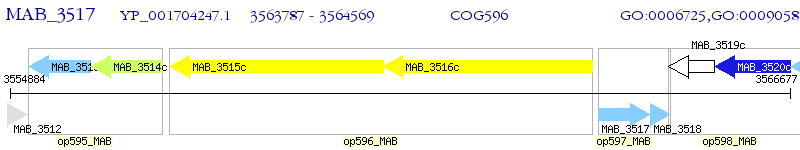
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3517 | - | - | 100% (260) | putative hydrolase, alpha/beta fold LipV |
| M. abscessus ATCC 19977 | MAB_3034 | - | 7e-12 | 35.58% (104) | hydrolase |
| M. abscessus ATCC 19977 | MAB_2946c | - | 4e-09 | 36.28% (113) | Alpha/beta hydrolase fold |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3228 | lipV | 2e-74 | 59.73% (221) | lipase LipV |
| M. gilvum PYR-GCK | Mflv_4667 | - | 3e-86 | 57.14% (259) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3203 | lipV | 2e-74 | 59.73% (221) | lipase LipV |
| M. leprae Br4923 | MLBr_01921 | - | 2e-05 | 33.33% (117) | hypothetical protein MLBr_01921 |
| M. marinum M | MMAR_1355 | lipV | 2e-79 | 55.00% (260) | lipase LipV |
| M. avium 104 | MAV_4150 | - | 4e-80 | 55.77% (260) | hydrolase, alpha/beta fold family protein |
| M. smegmatis MC2 155 | MSMEG_1940 | - | 3e-84 | 58.04% (255) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_0447 | lipV | 1e-86 | 58.50% (253) | POSSIBLE LIPASE LIPV |
| M. ulcerans Agy99 | MUL_2524 | lipV | 4e-80 | 55.38% (260) | lipase LipV |
| M. vanbaalenii PYR-1 | Mvan_1800 | - | 4e-84 | 55.60% (259) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3228|M.bovis_AF2122/97 --------------------------------------MPEIPIAAPDLL
Rv3203|M.tuberculosis_H37Rv --------------------------------------MPEIPIAAPDLL
MMAR_1355|M.marinum_M MSTDLHVHRYGPEEPVRVLTIHGVTEHGQSWEYLAG-YLPEVSIAAPDLL
MUL_2524|M.ulcerans_Agy99 MSTDLHVHRYGPEEPVRVLTIHGVTEHGQSWEYLAG-YLPEVNIAAPDLL
MAV_4150|M.avium_104 MTATLHVHRYGPPGPARILALHGLTGHGQRWQHLAG-LLPEFAWAAPDLV
TH_0447|M.thermoresistible__bu ------VYRYGPAGPPRLLAIHGLTGHGRRWQTLATRHLPEIAVAAPDLI
MSMEG_1940|M.smegmatis_MC2_155 ---MLHVHRYGPAGAAQVLLIHGLTGHGQRWQTLATEHLPEVGVLAPDLI
Mflv_4667|M.gilvum_PYR-GCK MTEQLCTYRFGPAGPARVLAIHGLTGHGKRWESLFTRHLSDIPAVAPDLV
Mvan_1800|M.vanbaalenii_PYR-1 MTDLLCTYRFGPPGPARVLAIHGLTGHGKRWETLFTRHLPDVTALAPDLL
MAB_3517|M.abscessus_ATCC_1997 MTPTLNIHRYGPVEPARLLALHGLTGHGRRWAPLFDEYLADVPVLAPDLL
MLBr_01921|M.leprae_Br4923 MSTNLLTLRGGRGKP--LILVHGLMGRGNTWSRQLPWLSQLGTVYTYDAP
: *
Mb3228|M.bovis_AF2122/97 GHGRSPWAAPWTIDAN--VSALAALLDNQGDGPVVVVGHSFGGAVAMHLA
Rv3203|M.tuberculosis_H37Rv GHGRSPWAAPWTIDAN--VSALAALLDNQGDGPVVVVGHSFGGAVAMHLA
MMAR_1355|M.marinum_M GHGRSSWAAPWSIDAN--VSALAALLKDQADGPVVVVGHSFGGAVALQLA
MUL_2524|M.ulcerans_Agy99 GHGRSSWAAPWSIDAN--VSALAALLKDQADGPVVVVGHSFGGAVALQLA
MAV_4150|M.avium_104 GHGRSSWSAPWTIDAN--ISALADLLDAQAGAPVLVVGHSFGAGLAMHLA
TH_0447|M.thermoresistible__bu GHGRSSWAAPWTFEAN--VGALVALLEDQADGPVVVVGHSFGGAVAMHLA
MSMEG_1940|M.smegmatis_MC2_155 GHGRSSWAAPWTIDAN--VAALAALLDR----PTLVVGHSFGGALALNLS
Mflv_4667|M.gilvum_PYR-GCK GHGRSSWDAPWTIEAN--VAALTALLDAEAEGPVVVVGHSFGGAIALNLA
Mvan_1800|M.vanbaalenii_PYR-1 GHGRSSWDAPWTIDAN--VAALAALVDAEADGPVVVVGHSFGGAIALNLA
MAB_3517|M.abscessus_ATCC_1997 GHGRSPATAPWSLEAH--ADAVAAELDTAETGPIVVVGHSFGGAIALHLA
MLBr_01921|M.leprae_Br4923 WHRGRNVVDPYPISTERFVTDLADAVSTLG-VPARLVGHSMGAMHSWCLA
* *:.:.:. :. :. * :****:*. : *:
Mb3228|M.bovis_AF2122/97 AARPDQVAALVLLDPAVALDGSRVREVVDAMLASPDYLDPAEARAEKATG
Rv3203|M.tuberculosis_H37Rv AARPDQVAALVLLDPAVALDGSRVREVVDAMLASPDYLDPAEARAEKATG
MMAR_1355|M.marinum_M AAHPDLVAALILLDPAIGLNGSRVREVVDAMVAFPDYTDPAEARMERAYG
MUL_2524|M.ulcerans_Agy99 AAHPDLVAALILLDPAIGLDGSRVREVVDAMVAFPDYTDPAEARMERAYG
MAV_4150|M.avium_104 AVRPDRVAGLLLLDPAIGLDGAWMREIAEAMLASPDYPDAEEARLEKLHG
TH_0447|M.thermoresistible__bu AARPDLVAGLVLLDPAIGLDGEWMREIAESMLASPDYPDREEARTEKFTG
MSMEG_1940|M.smegmatis_MC2_155 AAHPDLVSGLVLLDPAIGLDGEWMRDIADDMLSSPDYPDREEARLDKVNG
Mflv_4667|M.gilvum_PYR-GCK AARPDLIAGLVLLDPAVGLDGRWMREIADEMYTSPDYTDREEARQEKIGG
Mvan_1800|M.vanbaalenii_PYR-1 AVRPDLVAGLVLLDPAVALDGRWMREVADDMYNSPDYTDREEARQEKIGG
MAB_3517|M.abscessus_ATCC_1997 ARRPDLVKSLVLLDPAIGLDGDWMRQIADQMVTYPDYTDRAEAKSDKVAG
MLBr_01921|M.leprae_Br4923 AERPDLVSALVVEDMAPDFRGCSTGPWEPWLRGIPVEFDSVEKVFDEFGP
* :** : .*:: * * : * : * * * :.
Mb3228|M.bovis_AF2122/97 AWADVDPPVLDAELDEHLVALPNGRYGWRISLPAMVCYWSELARDIVLPP
Rv3203|M.tuberculosis_H37Rv AWADVDPPVLDAELDEHLVALPNGRYGWRISLPAMVCYWSELARDIVLPP
MMAR_1355|M.marinum_M AWADVDPAVLDAELDEHLITLPNGRYSWRISLPAMVCYWSELARDAALPP
MUL_2524|M.ulcerans_Agy99 AWADVDPAVLDAELDEHLITLPNGRYSWRISLPAMVCYWSELARDAALPP
MAV_4150|M.avium_104 SWSDVEPALLDAEIDEHLIELPNGRYGWRVCLPAMMSYWSELARDPVLPP
TH_0447|M.thermoresistible__bu SWADVEPAELDRDLDEHLIALPGGRFGWRICVPAMMAYWSELARDVVLPP
MSMEG_1940|M.smegmatis_MC2_155 SWGEVDPAEVERELDEHLVEVGEGRVGWRISIPAAMSYWSELARPVVLPH
Mflv_4667|M.gilvum_PYR-GCK SWGEVAEDELDRELDEHLVASAGGRVGWRISIPAMLCYWSELTRPAVIPR
Mvan_1800|M.vanbaalenii_PYR-1 SWGEVAEDELDRELDEHLVEQTDGRVGWRISIPATLSYWSELTRAVPVPR
MAB_3517|M.abscessus_ATCC_1997 AWSDVPDHVIEAELDEHLIQLSNGRCGWRMNLPAAVTAWSELARDIVLPV
MLBr_01921|M.leprae_Br4923 VAGQYFLDAFDRTSTGWQLHGHTSRWIEIAAEWGTRDYWEHWL------A
.: .: : .* . *..
Mb3228|M.bovis_AF2122/97 VGTATTLVRAVRASPAYVSDQLLAALDKRLGADFELLDFDCGHMVPQAKP
Rv3203|M.tuberculosis_H37Rv VGTATTLVRAVRASPAYVSDQLLAALDKRLGADFELLDFDCGHMVPQAKP
MMAR_1355|M.marinum_M PGIATTLVRAVRADPQYVSDELIAALRERLGADFALREFECGHMVPQAKP
MUL_2524|M.ulcerans_Agy99 PGIATTLVRAVRADPQYVSDELIAALRERLGADFALREFECGHMVPQAKP
MAV_4150|M.avium_104 AQTATILVRAKRTAPPYVGDELIAALQDRLGSRFRLLDFDCNHMVPYAKP
TH_0447|M.thermoresistible__bu SGIPTTLVRAAWTDPPYVTGELLDGLSQRLGADLTLVDLDCLHMVAQAKP
MSMEG_1940|M.smegmatis_MC2_155 NPIPTVLVRAKRCRPPFVADALIDGLTDRLGSKFTLLDWDCDHMVAQAKP
Mflv_4667|M.gilvum_PYR-GCK DGTPTTLVRATRTDPPYAGDELVAALDAGLGPNFTLLEWDCDHMVPLARP
Mvan_1800|M.vanbaalenii_PYR-1 DGTPTTLVRATHTDPPYASDELIATLDAGLGASLTVLEWDCDHMVPLAKP
MAB_3517|M.abscessus_ATCC_1997 NVIRTTLLLATRTSPPYVLPKLLDALQATLGSQFELIDIDCDHMVSQSRP
MLBr_01921|M.leprae_Br4923 VRSPVLLIEAGTSVTPHG---QMLAMTKTACRTTYLYVPEAGHLIHDEAT
. *: * . . : : : :. *:: .
Mb3228|M.bovis_AF2122/97 TEVAAVIRSRLGPR----
Rv3203|M.tuberculosis_H37Rv TEVAAVIRSRLGPR----
MMAR_1355|M.marinum_M TEVATLIREQLKTP----
MUL_2524|M.ulcerans_Agy99 TEVATLIREQLKTP----
MAV_4150|M.avium_104 DDVAALARELLKAR----
TH_0447|M.thermoresistible__bu AETAELIRRHLG------
MSMEG_1940|M.smegmatis_MC2_155 AETAELIRRQLG------
Mflv_4667|M.gilvum_PYR-GCK TETARLIRDLLP------
Mvan_1800|M.vanbaalenii_PYR-1 AETAAAIRDRLV------
MAB_3517|M.abscessus_ATCC_1997 AETAAAIRSHLE------
MLBr_01921|M.leprae_Br4923 QVYRQAVVSFLAGCAGRD
*