For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RKHVVLIHGAWSRGRQLAGARSAFEHHGYTVHTPTLRHHELPLPDGAARIATLSLRDYADDLVELVASLN SAPLLVGHSLGGLLVQLVAARTRHIGVVAACPSPVGPVGLNRTTAGLSMRHALRPRPWTKPVAPPSWPRF RTGIAGAQSAGAAREMFDDLVCESGRVLFFELALPWLDRSKVARVDYPAVTGPVLVLGGEYDRIVGSAIA RQTAGRYRNGTFVEIPGSDHLVFSARVLPATMARISEWIAHNHLFATA*
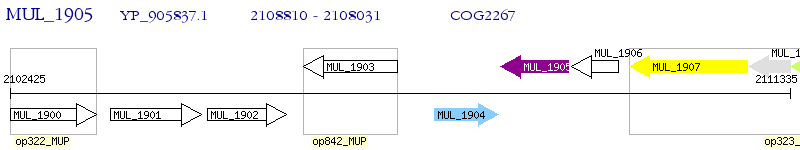
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1905 | - | - | 100% (259) | lysophospholipase |
| M. ulcerans Agy99 | MUL_4213 | - | 4e-06 | 27.20% (261) | hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3379 | - | 1e-08 | 28.35% (261) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1669 | - | 1e-148 | 98.46% (259) | lysophospholipase |
| M. avium 104 | MAV_3771 | - | 6e-11 | 26.59% (252) | hydrolase, alpha/beta fold family protein, putative |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2148 | - | 4e-10 | 28.84% (267) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3379|M.gilvum_PYR-GCK MAPAPLEVIDKGESTDAHPAPLLFVHGAWHAAWCWDDHFLDFFAGRGYRA
Mvan_2148|M.vanbaalenii_PYR-1 ----MLEVIDKGACSESHPVPLLFVHGAWHAAWCWDEHFLDFFADKGFRA
MAV_3771|M.avium_104 ----MLEVIEKGSGSAEHPTPLLFVHGGWHAAWCWEN-FLDFFADAGYRA
MUL_1905|M.ulcerans_Agy99 -----------------MRKHVVLIHGAWSRGRQLAG-ARSAFEHHGYTV
MMAR_1669|M.marinum_M -----------------MRKHVVLIHGAWSRGRQLAG-ARSAFEHHGYTV
::::**.* . . * *: .
Mflv_3379|M.gilvum_PYR-GCK LALSFRGHGRSATD--KPLRSLSISDYVDDVATVAARLPTPPIVVGHSMG
Mvan_2148|M.vanbaalenii_PYR-1 LAVSLRGHGNSPSP--KPLRTCSLADYLDDVDSVVDTLPTTPVLIGHSMG
MAV_3771|M.avium_104 VALSLRGHGASPTS--KPLHRVSIADYLDDVAAVAGELGGAPILIGHSLG
MUL_1905|M.ulcerans_Agy99 HTPTLRHHELPLPDGAARIATLSLRDYADDLVELVASLNSAPLLVGHSLG
MMAR_1669|M.marinum_M HTPTLRHHELPLPDGAARIATLSLRDYADDLVELVASLDSAPLLVGHSLG
: ::* * . . : *: ** **: :. * .*:::***:*
Mflv_3379|M.gilvum_PYR-GCK GFVVQKYLEDRDVPAGVLMASAPP------RGYLGSGVRWFRRHPWQFVR
Mvan_2148|M.vanbaalenii_PYR-1 GFIVQKYLEAHQAPAGVLIASAPQ------RGSFAFTMRLTRRHPWLTTK
MAV_3771|M.avium_104 GFVIQRYLETHRVPAAVLVGSVPP------QGVLRLALRVWRRRPSMTME
MUL_1905|M.ulcerans_Agy99 GLLVQLVAARTRHIGVVAACPSPVGPVGLNRTTAGLSMRHALR-PRPWTK
MMAR_1669|M.marinum_M GLLVQLVAARTRHIGVVAACPSPVGPVGLNRTTAGLSMRHALR-PRPWTK
*:::* . * . * : :* * * .
Mflv_3379|M.gilvum_PYR-GCK TSLSGESLAYVSPIEAARERFFSPSTPEAIVAACAARLQEESARSGR-DG
Mvan_2148|M.vanbaalenii_PYR-1 GLITGNALLPVGTPELARESFFSSQTPESDVVRYAARLNGESQRVAI-DT
MAV_3771|M.avium_104 AWNDPTLLKFLATPALAREYLFCAATPEAIVESCRQRAGAESVRAAMTDP
MUL_1905|M.ulcerans_Agy99 PVAPPSWPRFRTGIAGAQSAGAAREMFDDLVCESGRVLFFELALPWLDRS
MMAR_1669|M.marinum_M PVAPPSWPRFRTGIAGAQSAGAAREMFEDLVCESGRVLFFELALPWLDRS
*:. . : * *
Mflv_3379|M.gilvum_PYR-GCK VTALPRPKRVRAPMLVLGALDDGMAVTPAEVHATARAYRT-EATLFPGMG
Mvan_2148|M.vanbaalenii_PYR-1 IWMLPRPKRVTTPLLILGAECD-RSILSKEVRTTANAYRT-EAEIFPDMG
MAV_3771|M.avium_104 MLRRVRTRRVSTPILVLGATHD-GFVSAADVRATARAYRT-DPEFF-DMG
MUL_1905|M.ulcerans_Agy99 KVARVDYPAVTGPVLVLGGEYD-RIVGSAIARQTAGRYRNGTFVEIPGSD
MMAR_1669|M.marinum_M KVARVDYPAVTGPVLVLGGEYD-RIVGSTIARQTAGRYRNGTFVEIPGSD
* *:*:**. * : . .: ** **. : . .
Mflv_3379|M.gilvum_PYR-GCK HNMMLEPGWEAVASHIDSWLISRGL----------------
Mvan_2148|M.vanbaalenii_PYR-1 HDMMLEPGWAAVAERIHTWLEMRDLRPHRGEPDQQETTPTG
MAV_3771|M.avium_104 HNMMLEPGWVAVAERIRDWLQAPTAARHR------------
MUL_1905|M.ulcerans_Agy99 HLVFSARVLPATMARISEWIAHNHLFATA------------
MMAR_1669|M.marinum_M HLVFSARVLPATMARISEWIAHNQLFATA------------
* :: *. :* *: