For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PMHNRLLGKGVLVTGAASGIGEATVRRLLDEGASVVGCDLAPGPPSDLPGDIGYLSGDVRDTATAERAVA AVIERTGRLDGVVHSAGVGGGGPIHLLPDAEWDRVLDVNLKGTFVVLRAALAQMITQDRVDGERGAIVTL SSIEGLEGTAGGSAYNASKGGVVLLTKNAAIDYGPSGIRVNAICPGFIETPLFESVIGIPGMEGPRDGLR HEHKLRRFGRPEEVAAVAAFLVSSDAAFVSGQAIAVDGGYTAGRDHHVTELMGLGGP*
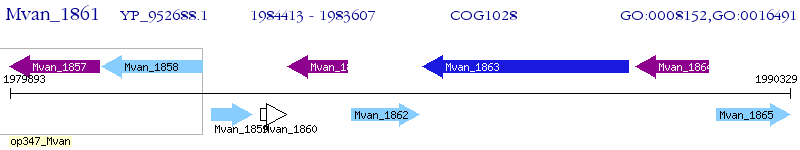
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1861 | - | - | 100% (268) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_3000 | - | 3e-35 | 37.69% (260) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_2281 | - | 3e-34 | 35.36% (263) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 5e-36 | 36.92% (260) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4508 | - | 1e-126 | 85.00% (260) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 5e-36 | 36.92% (260) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 4e-27 | 31.98% (247) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4680c | - | 1e-32 | 35.83% (254) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_3470 | - | 3e-93 | 68.50% (254) | short chain membrane-associated dehydrogenase |
| M. avium 104 | MAV_3521 | - | 1e-92 | 66.28% (261) | NAD dependent epimerase/dehydratase family protein |
| M. smegmatis MC2 155 | MSMEG_3009 | - | 8e-37 | 37.01% (254) | 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase |
| M. thermoresistible (build 8) | TH_4254 | - | 1e-39 | 42.02% (257) | PUTATIVE putative 2,5-dichloro-2,5-cyclohexadiene-1,4-diol |
| M. ulcerans Agy99 | MUL_0315 | fabG2 | 1e-35 | 38.66% (238) | 3-oxoacyl- |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1861|M.vanbaalenii_PYR-1 ------------MPMHNRLLGKGVLVTGAASGIGEATVRRLLDEGASVVG
Mflv_4508|M.gilvum_PYR-GCK -------MWAILWRVVKGLAGKGVLVTGAASGIGLATVRRLLEEGAAVVG
MMAR_3470|M.marinum_M ------------ML--DHRASRTFVVTGASSGIGLATVGRLLEQGCAVVG
MAV_3521|M.avium_104 ---------MTGMVNPDRARSRTFVVTGAASGIGLATARRLLAEGGTVVG
MSMEG_3009|M.smegmatis_MC2_155 ---------------MNRLDGKVAIVTGTCRGIGHAIADALAREGAITVA
TH_4254|M.thermoresistible__bu -----------MTAGSTRLAGKVVLVTGAGGGLGTAIARRLADEGARLVL
Mb2025|M.bovis_AF2122/97 -----------MSGR---LIGKVALVSGGARGMGASHVRAMVAEGAKVVF
Rv2002|M.tuberculosis_H37Rv -----------MSGR---LIGKVALVSGGARGMGASHVRAMVAEGAKVVF
MAB_4680c|M.abscessus_ATCC_199 -----------MNGNSALLQGKNVIVTGGARGLGAAFSRHIVSQGGRVVI
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
MUL_0315|M.ulcerans_Agy99 ---------MRPD-SSPSWCTITSVVTGGASGIGLAIARRLSADGTDVAT
:*:* *:* : : :* .
Mvan_1861|M.vanbaalenii_PYR-1 CDLAPG--------PPSDLPGDIGYLSGDVRDTATAERAVAAVIERTGRL
Mflv_4508|M.gilvum_PYR-GCK MDVCTE--------APADVPGDFTYHRGDVLDAAAVGRAVTAVVERTGRL
MMAR_3470|M.marinum_M ADVVT---------PPHDLGPRFTFVTADVTDEDAVAGVFDAVPD---RL
MAV_3521|M.avium_104 VDLAD---------PPGDLGPRFTFVPADVTDESAVAAVLAAVPG---RL
MSMEG_3009|M.smegmatis_MC2_155 TGRSKT------DEALGPRSAEVEYRQLDVASEEDWDSVVTETVERYGRI
TH_4254|M.thermoresistible__bu ADLQESQCAALADELPAPAGAHHPLG-FDVADETAWISAISTVSAQYGAL
Mb2025|M.bovis_AF2122/97 GDILDEE----GKAVAAELADAARYVHLDVTQPAQWTAAVDTAVTAFGGL
Rv2002|M.tuberculosis_H37Rv GDILDEE----GKAVAAELADAARYVHLDVTQPAQWTAAVDTAVTAFGGL
MAB_4680c|M.abscessus_ATCC_199 ADVLDGD----GRQLAAELGDAARFTHLDVTDAEQWRELIDTTLAGFGTI
MLBr_01807|M.leprae_Br4923 THRASV------------VPDWFFGVECDVTDNDAIGAAFKAVEEHQGPV
MUL_0315|M.ulcerans_Agy99 LDLK--------------TPDGGLAFSADVTNRSQVDAALAAIRQRLGPV
** . . :
Mvan_1861|M.vanbaalenii_PYR-1 DGVVHSAGVGGGGPIHLLPDAEWDRVLDVNLKGTFVVLRAALAQMITQDR
Mflv_4508|M.gilvum_PYR-GCK DGVVHSAGVGGGGPIHLLPDDEWDRVVDVNLKGTFVVLRAALAQMTTQDR
MMAR_3470|M.marinum_M DGVVHSAGVAGGGPVHLLARAEWDRVIGVNLTGTFLVAKAALARMIEQPR
MAV_3521|M.avium_104 DGVFHAAGVAGGGPVHLLDRSEWDRVIGVNLTGTFLVAKAALARMIDQPR
MSMEG_3009|M.smegmatis_MC2_155 DVLVNNAGIIAYQGLHELTVPEWNRVVATNQTGTWLGMRAVIPHMVSQ--
TH_4254|M.thermoresistible__bu HALVNNAAIGALGNVAEERLERWDRILDVGATGVWLGMKHAVPLIRRS--
Mb2025|M.bovis_AF2122/97 HVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEA--
Rv2002|M.tuberculosis_H37Rv HVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEA--
MAB_4680c|M.abscessus_ATCC_199 TGLVNNAGISSAAMVADEPLEHFRKVIEINLVGVFIGMQAVIPAMRAH--
MLBr_01807|M.leprae_Br4923 EVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR-
MUL_0315|M.ulcerans_Agy99 TILVNAAGLDGFKRFSNITFEDWQRVIDVNLNGVFHTIQAVLPDMVAAG-
:. *.: . : ::: . *.: : . :
Mvan_1861|M.vanbaalenii_PYR-1 VDGERGAIVTLSSIEGLEGTAG-GSAYNASKGGVVLLTKNAAIDYGPSGI
Mflv_4508|M.gilvum_PYR-GCK VTGERGSIVTLSSVEGLEGTAG-GSAYNASKGGVVLLTKNAAIDYGPSGI
MMAR_3470|M.marinum_M VDGERGSIVTLASIEGLEGTAG-GSSYNAAKGGVVLLTKNIALDYGPSGI
MAV_3521|M.avium_104 VDGERGSLVTVASVEGLEGTAG-GSSYNAAKGGVVLLTKNIALDYGPSGI
MSMEG_3009|M.smegmatis_MC2_155 ---HGGSIINISSIWGSAAVAG-AHAYHATKGAVRNMSKSAAISYAKDNV
TH_4254|M.thermoresistible__bu ---GGGSVVNICSIFGTVGGFGDSAAYHAAKGAVRTLTKNAALHWAPDGV
Mb2025|M.bovis_AF2122/97 ---GRGSIINISSIEGLAGTVA-CHGYTATKFAVRGLTKSTALELGPSGI
Rv2002|M.tuberculosis_H37Rv ---GRGSIINISSIEGLAGTVA-CHGYTATKFAVRGLTKSTALELGPSGI
MAB_4680c|M.abscessus_ATCC_199 ---GCGSIVNISSAAGLVGLAL-TSGYGASKWGVRGLSKVAAIELGRERI
MLBr_01807|M.leprae_Br4923 ----FGRYIFMGSVVGLSGVPG-QANYAASKSGLIGMARSLARELGSRSI
MUL_0315|M.ulcerans_Agy99 ----WGRIVNISSSSTHSGTPY-MTHYVAAKSAVNGLTKALALEYGPSGI
* : : * . * *:* .: ::: * . :
Mvan_1861|M.vanbaalenii_PYR-1 RVNAICPGFIETPLFESVIGIPGMEGPRDGLRHEHKLRRFGRPEEVAAVA
Mflv_4508|M.gilvum_PYR-GCK RINAICPGFIETPLFDSVMGLPGMEGPRDGLRHEHKLRRFGRPEEVASVA
MMAR_3470|M.marinum_M RANAICPGFIATPLAEGVFGMPGMEGPRASITNEHALQRLGKPEEIAAMA
MAV_3521|M.avium_104 RANVICPGFIETPMAHNVFSIPGMEAPLASITREHALQRLGRPEEIAAMA
MSMEG_3009|M.smegmatis_MC2_155 RVNSVHPGFIRTP-LTDAQETTIN----DYVVSQTPMGRAGTPDEIANGV
TH_4254|M.thermoresistible__bu RVNSLHPGFIETPQLRELYKGTERY---ERMLAGTPLGRLGRPDEIAAVV
Mb2025|M.bovis_AF2122/97 RVNSIHPGLVKTP--MTDWV--------PEDIFQTALGRAAEPVEVSNLV
Rv2002|M.tuberculosis_H37Rv RVNSIHPGLVKTP--MTDWV--------PEDIFQTALGRAAEPVEVSNLV
MAB_4680c|M.abscessus_ATCC_199 RVNSVHPGVIYTP--MTAKEGFREG---EGNMPIAAMGRVGNADEVAGAV
MLBr_01807|M.leprae_Br4923 TANVVAPGFIDTEMTRAMDSK-----YQEAALQAIPLGRIGAVEEIAGAV
MUL_0315|M.ulcerans_Agy99 TVNAVPPGFIDTPMLRNAAAQGHLG-DIEQNIARTPVRRIGKPEDIAAAC
* : **.: * : * . :::
Mvan_1861|M.vanbaalenii_PYR-1 AFLVSSDAAFVSGQAIAVDGGYTAGRDHHVTELMGLGGP-----
Mflv_4508|M.gilvum_PYR-GCK TFLVSAEASFVSGQAIAVDGGYTAGRDHHVTELLGLGEQ-----
MMAR_3470|M.marinum_M AFLLSPDASFVTGQAIAVDGGYTAGRDHGVVRLFGLPQ------
MAV_3521|M.avium_104 AFLLSTDASFVSGQAIAVDGGYTAGRDHGVVELFGFPS------
MSMEG_3009|M.smegmatis_MC2_155 VFLASDEASFVTGSELVIDGGYLAQ-------------------
TH_4254|M.thermoresistible__bu AFLVSDDASFITGSEVYADGGWTAR-------------------
Mb2025|M.bovis_AF2122/97 VYLASDESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
Rv2002|M.tuberculosis_H37Rv VYLASDESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
MAB_4680c|M.abscessus_ATCC_199 AYLLSDAASYVTGAELAVDGGWTAGPT-----LTTITGQ-----
MLBr_01807|M.leprae_Br4923 SFLASEDAGYISGAVIPVDGGLGMGH------------------
MUL_0315|M.ulcerans_Agy99 AFLVSEEAGYITGQILGVNGGRNT--------------------
:* * :.: :* . :**