For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LDHRASRTFVVTGASSGIGLATVGRLLEQGCAVVGADVVTPPHDLGPRFTFVTADVTDEDAVAGVFDAVP DRLDGVVHSAGVAGGGPVHLLARAEWDRVIGVNLTGTFLVAKAALARMIEQPRVDGERGSIVTLASIEGL EGTAGGSSYNAAKGGVVLLTKNIALDYGPSGIRANAICPGFIATPLAEGVFGMPGMEGPRASITNEHALQ RLGKPEEIAAMAAFLLSPDASFVTGQAIAVDGGYTAGRDHGVVRLFGLPQ*
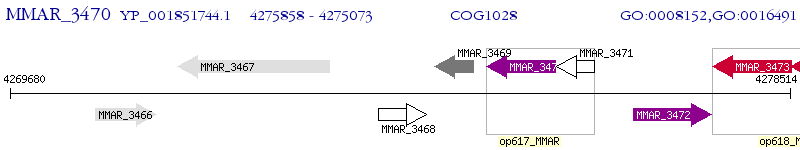
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3470 | - | - | 100% (261) | short chain membrane-associated dehydrogenase |
| M. marinum M | MMAR_4715 | - | 2e-34 | 39.17% (240) | 3-oxoacyl- |
| M. marinum M | MMAR_2981 | fabG3_1 | 1e-32 | 33.73% (249) | 20-beta-hydroxysteroid dehydrogenase FabG3_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 5e-34 | 36.65% (251) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4508 | - | 9e-93 | 67.05% (258) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2857c | - | 6e-34 | 36.65% (251) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-31 | 35.46% (251) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0827 | - | 6e-29 | 35.29% (255) | putative short-chain dehydrogenase/reductase |
| M. avium 104 | MAV_3521 | - | 1e-120 | 82.95% (258) | NAD dependent epimerase/dehydratase family protein |
| M. smegmatis MC2 155 | MSMEG_3830 | - | 2e-34 | 39.11% (248) | 3-oxoacyl- |
| M. thermoresistible (build 8) | TH_3102 | - | 2e-32 | 38.82% (237) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0315 | fabG2 | 6e-34 | 39.33% (239) | 3-oxoacyl- |
| M. vanbaalenii PYR-1 | Mvan_1861 | - | 3e-93 | 68.50% (254) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3470|M.marinum_M -----ML--DHRAS------------RTFVVTGASSGIGLATVGRLLEQG
MAV_3521|M.avium_104 --MTGMVNPDRARS------------RTFVVTGAASGIGLATARRLLAEG
Mflv_4508|M.gilvum_PYR-GCK MWAILWRVVKGLAG------------KGVLVTGAASGIGLATVRRLLEEG
Mvan_1861|M.vanbaalenii_PYR-1 -----MPMHNRLLG------------KGVLVTGAASGIGEATVRRLLDEG
TH_3102|M.thermoresistible__bu -------------------------LKNAVVTGGGSGIGQAVAARLRADG
MUL_0315|M.ulcerans_Agy99 -----MRPDSS----------PSWCTITSVVTGGASGIGLAIARRLSADG
MLBr_01807|M.leprae_Br4923 -----MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADG
MAB_0827|M.abscessus_ATCC_1997 -----MTSPLK--------------SRRAFVTGGSRGIGAGIVRRLAEDG
Mb2882c|M.bovis_AF2122/97 -----MMDLSQR-----------LAGRVAVITGGGSGIGLAAGRRMRAEG
Rv2857c|M.tuberculosis_H37Rv -----MMDLSQR-----------LAGRVAVITGGGSGIGLAAGRRMRAEG
MSMEG_3830|M.smegmatis_MC2_155 ------------------------MSRTALVTGANGGIGSATVARLVKNG
.:**. *** . *: :*
MMAR_3470|M.marinum_M CAVVGADVVT-PPHDLG--------PRFTFVTADVTDEDAVAGVFDAVPD
MAV_3521|M.avium_104 GTVVGVDLAD-PPGDLG--------PRFTFVPADVTDESAVAAVLAAVPG
Mflv_4508|M.gilvum_PYR-GCK AAVVGMDVCTEAPADVP--------GDFTYHRGDVLDAAAVGRAVTAVVE
Mvan_1861|M.vanbaalenii_PYR-1 ASVVGCDLAPGPPSDLP--------GDIGYLSGDVRDTATAERAVAAVIE
TH_3102|M.thermoresistible__bu YHVATIDLKPGDD--------------EFSHAADVTDRAQVDAAFDAIRT
MUL_0315|M.ulcerans_Agy99 TDVATLDLKTPDG--------------GLAFSADVTNRSQVDAALAAIRQ
MLBr_01807|M.leprae_Br4923 HRVAVTHRASVVP------------DWFFGVECDVTDNDAIGAAFKAVEE
MAB_0827|M.abscessus_ATCC_1997 ARVVFTYSSSVQQAADVVDDINEDGGWARAVKADVADPDQLGAAIGQAAE
Mb2882c|M.bovis_AF2122/97 ATIVVGDVDVEAGG------AAADELSGLFVPTDVCDEDAVNGLFDGAAE
Rv2857c|M.tuberculosis_H37Rv ATIVVGDVDVEAGG------AAADELSGLFVPTDVCDEDAVNGLFDGAAE
MSMEG_3830|M.smegmatis_MC2_155 VEVIATDLGERSD--------------TVPADADYVPFDLRSGDPHELFE
: *
MMAR_3470|M.marinum_M -----RLDGVVHSAGVAG---GGPVHLLARAEWDRVIGVNLTGTFLVAKA
MAV_3521|M.avium_104 -----RLDGVFHAAGVAG---GGPVHLLDRSEWDRVIGVNLTGTFLVAKA
Mflv_4508|M.gilvum_PYR-GCK RTG--RLDGVVHSAGVGG---GGPIHLLPDDEWDRVVDVNLKGTFVVLRA
Mvan_1861|M.vanbaalenii_PYR-1 RTG--RLDGVVHSAGVGG---GGPIHLLPDAEWDRVLDVNLKGTFVVLRA
TH_3102|M.thermoresistible__bu QLG--PVTILVNAAGLDL---FRRFTDISFEDWERVINVNLHGVFHCIQA
MUL_0315|M.ulcerans_Agy99 RLG--PVTILVNAAGLDG---FKRFSNITFEDWQRVIDVNLNGVFHTIQA
MLBr_01807|M.leprae_Br4923 HQG--PVEVLVANAGITS---DMLIMRMSEEQFQKVIDVNLTGVFRVAKL
MAB_0827|M.abscessus_ATCC_1997 ELG--GLDILVNNAGILS---IGDVDTATLADFDEIVAVNVRAVFAGIKA
Mb2882c|M.bovis_AF2122/97 TYG--RIDIAFNNAGISPPE-DNLIENTELAAWQRVQDVNLKSVYLCCRA
Rv2857c|M.tuberculosis_H37Rv TYG--RIDIAFNNAGISPPE-DNLIENTELAAWQRVQDVNLKSVYLCCRA
MSMEG_3830|M.smegmatis_MC2_155 RLGGRGLDYLVNAAGVALFDRDGSMLDIDESIWDVTLGVNLHGLRKVTAA
: . **: . :: **: .
MMAR_3470|M.marinum_M ALARMIEQPRVDGERGSIVTLASIEG-LEGTAGGSSYNAAKGGVVLLTKN
MAV_3521|M.avium_104 ALARMIDQPRVDGERGSLVTVASVEG-LEGTAGGSSYNAAKGGVVLLTKN
Mflv_4508|M.gilvum_PYR-GCK ALAQMTTQDRVTGERGSIVTLSSVEG-LEGTAGGSAYNASKGGVVLLTKN
Mvan_1861|M.vanbaalenii_PYR-1 ALAQMITQDRVDGERGAIVTLSSIEG-LEGTAGGSAYNASKGGVVLLTKN
TH_3102|M.thermoresistible__bu VLPDMIEAG-----WGRIVNISSSST-HSGAPYMAPYVAAKSGVNGLTKT
MUL_0315|M.ulcerans_Agy99 VLPDMVAAG-----WGRIVNISSSST-HSGTPYMTHYVAAKSAVNGLTKA
MLBr_01807|M.leprae_Br4923 ASRNMIKQR-----FGRYIFMGSVVG-LSGVPGQANYAASKSGLIGMARS
MAB_0827|M.abscessus_ATCC_1997 ASPHLTEG-------GRIVTIGSINGDSSPTANLSLYSMTKAAVAGLTRG
Mb2882c|M.bovis_AF2122/97 ALRHMVLAG-----KGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRE
Rv2857c|M.tuberculosis_H37Rv ALRHMVLAG-----KGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRE
MSMEG_3830|M.smegmatis_MC2_155 AVEHLRRGS-----GKSIVNVASTAGLRGMDSPLDAYQVSKAAVVSLTRA
. : : .* . * :*..: :::
MMAR_3470|M.marinum_M IALDYGPSGIRANAICPGFIATPLAEGVFGMPGMEGPRASITNEHALQRL
MAV_3521|M.avium_104 IALDYGPSGIRANVICPGFIETPMAHNVFSIPGMEAPLASITREHALQRL
Mflv_4508|M.gilvum_PYR-GCK AAIDYGPSGIRINAICPGFIETPLFDSVMGLPGMEGPRDGLRHEHKLRRF
Mvan_1861|M.vanbaalenii_PYR-1 AAIDYGPSGIRVNAICPGFIETPLFESVIGIPGMEGPRDGLRHEHKLRRF
TH_3102|M.thermoresistible__bu LALEYGPAGITVNAVPPGFIDTPMLRAAAAKG-YLGDIDKTIEATPVRRM
MUL_0315|M.ulcerans_Agy99 LALEYGPSGITVNAVPPGFIDTPMLRNAAAQG-HLGDIEQNIARTPVRRI
MLBr_01807|M.leprae_Br4923 LARELGSRSITANVVAPGFIDTEMTRAMDSK-----YQEAALQAIPLGRI
MAB_0827|M.abscessus_ATCC_1997 LAREFGPRGITINNVQPGPIGTDMN-PEDGQ-----LADILRPNIAVGRY
Mb2882c|M.bovis_AF2122/97 LGVQFARQGIRVNALCPGPVNTPLLQELFAKN--PERAARRMVHVPLGRF
Rv2857c|M.tuberculosis_H37Rv LGVQFARQGIRVNALCPGPVNTPLLQELFAKN--PERAARRMVHVPLGRF
MSMEG_3830|M.smegmatis_MC2_155 LALQLAPEGIRANTVCPGAILTPMIDHLYVEN--PARRVDMENRTPMRRL
. : . .* * : ** : * : : *
MMAR_3470|M.marinum_M GKPEEIAAMAAFLLSPDASFVTGQAIAVDGGYTAGRDHGVVRLFGLPQ-
MAV_3521|M.avium_104 GRPEEIAAMAAFLLSTDASFVSGQAIAVDGGYTAGRDHGVVELFGFPS-
Mflv_4508|M.gilvum_PYR-GCK GRPEEVASVATFLVSAEASFVSGQAIAVDGGYTAGRDHHVTELLGLGEQ
Mvan_1861|M.vanbaalenii_PYR-1 GRPEEVAAVAAFLVSSDAAFVSGQAIAVDGGYTAGRDHHVTELMGLGGP
TH_3102|M.thermoresistible__bu GKPEDIAAACAFLISEEAGYITGQILGVNGGRNT---------------
MUL_0315|M.ulcerans_Agy99 GKPEDIAAACAFLVSEEAGYITGQILGVNGGRNT---------------
MLBr_01807|M.leprae_Br4923 GAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH-------------
MAB_0827|M.abscessus_ATCC_1997 GQPRDIASLVSYLVSDEAWYITGTAINIDGGFTA---------------
Mb2882c|M.bovis_AF2122/97 AEPDEIAAAVAFLASDDASFITASTFLVDGGISSAYVTPL---------
Rv2857c|M.tuberculosis_H37Rv AEPDEIAAAVAFLASDDASFITASTFLVDGGISSAYVTPL---------
MSMEG_3830|M.smegmatis_MC2_155 GMPDEIASAIAFLLSDEASFVTATDLVVDGGWTAQIK------------
. ::*. ::* * :* :::. : ::**