For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GRVDGKVALISGGAQGMGAADARALIAEGAKVVIGDILDDKGKALADEINAETPDSIRYVHLDVTQADQW EAAVATAVDAFGKLNVLVNNAGTVALGQIGQFDMAKWQKVIDVNLTGTFLGMQASVEAMKTAGGGSIINI SSIEGLRGAVMVHPYVASKWAVRGLTKSAALELGSHNIRVNSVHPGFIRTPMTKHFPDNMLRIPLGRPGQ PEEVATFVVFLASDESRYATGAEFVMDGGLTNDVPHK*
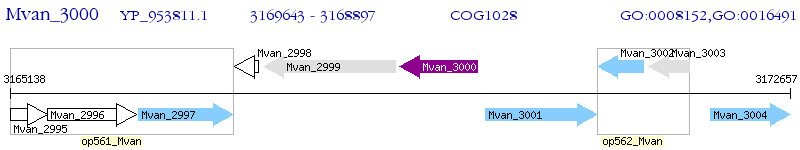
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3000 | - | - | 100% (248) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_1975 | fabG | 6e-45 | 40.83% (240) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_4237 | - | 3e-41 | 40.54% (259) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 2e-82 | 61.79% (246) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3281 | - | 1e-134 | 95.16% (248) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 2e-82 | 61.79% (246) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 3e-30 | 37.97% (237) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4680c | - | 6e-58 | 47.13% (244) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0387 | fabG3 | 1e-107 | 77.82% (248) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. avium 104 | MAV_5136 | - | 1e-110 | 79.84% (248) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3515 | - | 1e-114 | 82.66% (248) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. thermoresistible (build 8) | TH_4254 | - | 5e-47 | 40.73% (248) | PUTATIVE putative 2,5-dichloro-2,5-cyclohexadiene-1,4-diol |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 1e-107 | 77.42% (248) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2025|M.bovis_AF2122/97 --------------MSGRLIGKVALVSGGARGMGASHVRAMVAEGAKVVF
Rv2002|M.tuberculosis_H37Rv --------------MSGRLIGKVALVSGGARGMGASHVRAMVAEGAKVVF
Mvan_3000|M.vanbaalenii_PYR-1 ---------------MGRVDGKVALISGGAQGMGAADARALIAEGAKVVI
Mflv_3281|M.gilvum_PYR-GCK ---------------MGRVDGKVALISGGAQGMGAEDARALIAEGAKVVI
MSMEG_3515|M.smegmatis_MC2_155 ---------------MGRVAGKVALISGGARGMGAAHARALVAEGAKVVI
MMAR_0387|M.marinum_M ---------------MGRVDGKVALISGGARGMGASHARLLVQEGAKVVI
MUL_1037|M.ulcerans_Agy99 ---------------MGRVDGKVALISGGARGMGASHARLLVQEGAKVVI
MAV_5136|M.avium_104 ---------------MGRVDGKVALISGGARGMGAEHARLLAAEGAKVVI
MAB_4680c|M.abscessus_ATCC_199 -----------MNGNSALLQGKNVIVTGGARGLGAAFSRHIVSQGGRVVI
TH_4254|M.thermoresistible__bu -----------MTAGSTRLAGKVVLVTGAGGGLGTAIARRLADEGARLVL
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
. .: :::*. *:* * : :* ::..
Mb2025|M.bovis_AF2122/97 GDILDEEGKAVAAELA----DAARYVH---LDVTQPAQWTAAVDTAVTAF
Rv2002|M.tuberculosis_H37Rv GDILDEEGKAVAAELA----DAARYVH---LDVTQPAQWTAAVDTAVTAF
Mvan_3000|M.vanbaalenii_PYR-1 GDILDDKGKALADEINAETPDSIRYVH---LDVTQADQWEAAVATAVDAF
Mflv_3281|M.gilvum_PYR-GCK GDILDEKGQALADEINAQTPDSIRYVH---LDVTQADQWEAAVATAVNDF
MSMEG_3515|M.smegmatis_MC2_155 GDILDDEGAALAAELG----DAARYVH---LDVTQAEDWDTAVATATTDF
MMAR_0387|M.marinum_M GDILDEEGKALAEEIG----DAARYVH---LDVTQPDQWEAAVATAVDEF
MUL_1037|M.ulcerans_Agy99 GDILDEEGKALAEEIG----DAARYVH---LDVTQPDQWEAAVATAVDEF
MAV_5136|M.avium_104 GDILDDEGKAVADEIG----DSVRYVH---LDVTQPDQWDAAVETAVGEF
MAB_4680c|M.abscessus_ATCC_199 ADVLDGDGRQLAAELG----DAARFTH---LDVTDAEQWRELIDTTLAGF
TH_4254|M.thermoresistible__bu ADLQESQCAALADELP----APAGAHHPLGFDVADETAWISAISTVSAQY
MLBr_01807|M.leprae_Br4923 THRASVVPDWFFGVEC---------------DVTDNDAIGAAFKAVEEHQ
. . . **:: . :.
Mb2025|M.bovis_AF2122/97 GGLHVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKE
Rv2002|M.tuberculosis_H37Rv GGLHVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKE
Mvan_3000|M.vanbaalenii_PYR-1 GKLNVLVNNAGTVALGQIGQFDMAKWQKVIDVNLTGTFLGMQASVEAMKT
Mflv_3281|M.gilvum_PYR-GCK GTLNVLVNNAGTVALGQIGQFDMAKWQKVIDVNLTGTFLGMQASVEAMKA
MSMEG_3515|M.smegmatis_MC2_155 GLLNVLINNAGIVALGAIGKFDMTKWQNVIDVNLTGTFLGMQASVGAMKA
MMAR_0387|M.marinum_M GKLDVLVNNAGIVALGQLKKFDLGKWQKVIDVNLTGTFLGMRAAVEPMTA
MUL_1037|M.ulcerans_Agy99 GKLDVLVNNVGIVALGQLKKFDLGKWQKVIDVNLTGTFLGMRAAVEPMTA
MAV_5136|M.avium_104 GKLNVLVNNAGTVALGPLKSFDLAKWQKVIDVNLTGTFLGMRVAVEPMIA
MAB_4680c|M.abscessus_ATCC_199 GTITGLVNNAGISSAAMVADEPLEHFRKVIEINLVGVFIGMQAVIPAMRA
TH_4254|M.thermoresistible__bu GALHALVNNAAIGALGNVAEERLERWDRILDVGATGVWLGMKHAVPLIRR
MLBr_01807|M.leprae_Br4923 GPVEVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIK
* : *: *.. : .: .::::. .*.: : :
Mb2025|M.bovis_AF2122/97 AGRGSIINISSIEGLAG-TVACHGYTATKFAVRGLTKSTALELGPSGIRV
Rv2002|M.tuberculosis_H37Rv AGRGSIINISSIEGLAG-TVACHGYTATKFAVRGLTKSTALELGPSGIRV
Mvan_3000|M.vanbaalenii_PYR-1 AGGGSIINISSIEGLRG-AVMVHPYVASKWAVRGLTKSAALELGSHNIRV
Mflv_3281|M.gilvum_PYR-GCK AGGGSIINISSIEGLRG-AIMVHPYVASKWAVRGLTKSAALELGQYNIRV
MSMEG_3515|M.smegmatis_MC2_155 AGGGSIINVSSIEGLRG-APMVHPYVASKWAVRGLTKSAALELGAHQIRV
MMAR_0387|M.marinum_M AGSGSIINVSSIEGLRG-APAVHPYVASKWAVRGLTKSAALELAPLNIRV
MUL_1037|M.ulcerans_Agy99 AGSGSIINVSSIEGLRG-APAVHPYVASKWAVRGLTKSAALELAPLNIRV
MAV_5136|M.avium_104 AGGGSIINISSIEGLRG-APMVHPYVASKWGVRGLAKSAALELAPHNIRV
MAB_4680c|M.abscessus_ATCC_199 HGCGSIVNISSAAGLVG-LALTSGYGASKWGVRGLSKVAAIELGRERIRV
TH_4254|M.thermoresistible__bu SGGGSVVNICSIFGTVGGFGDSAAYHAAKGAVRTLTKNAALHWAPDGVRV
MLBr_01807|M.leprae_Br4923 QRFGRYIFMGSVVGLSG-VPGQANYAASKSGLIGMARSLARELGSRSITA
* : : * * * * *:* .: ::: * . . : .
Mb2025|M.bovis_AF2122/97 NSIHPGLVKTPMT-------DWVPEDIFQTALGRAAEPVEVSNLVVYLAS
Rv2002|M.tuberculosis_H37Rv NSIHPGLVKTPMT-------DWVPEDIFQTALGRAAEPVEVSNLVVYLAS
Mvan_3000|M.vanbaalenii_PYR-1 NSVHPGFIRTPMT-------KHFPDNMLRIPLGRPGQPEEVATFVVFLAS
Mflv_3281|M.gilvum_PYR-GCK NSVHPGFIRTPMT-------KHFPDNMLRIPLGRPGQPEEVATFVVFLAS
MSMEG_3515|M.smegmatis_MC2_155 NSIHPGFIRTPMT-------EHFPDDMLRIPLGRPGEVDEVSSFVVFLAS
MMAR_0387|M.marinum_M NSIHPGFIRTPMT-------ANLPDDMVTIPLGRPAESREVSTFVVFLAS
MUL_1037|M.ulcerans_Agy99 NSIHPGFIRTPMT-------ANLPDDMVTIPLGRPAESREVSTFVVFLAS
MAV_5136|M.avium_104 NSVHPGFIRTPMT-------KHLPDDMVTVPLGRPAESREVSTFVLFLAS
MAB_4680c|M.abscessus_ATCC_199 NSVHPGVIYTPMT--AKEGFREGEGNMPIAAMGRVGNADEVAGAVAYLLS
TH_4254|M.thermoresistible__bu NSLHPGFIETPQLRELYKGTERYERMLAGTPLGRLGRPDEIAAVVAFLVS
MLBr_01807|M.leprae_Br4923 NVVAPGFIDTEMTR--AMDSKYQEAALQAIPLGRIGAVEEIAGAVSFLAS
* : **.: * : .:** . *:: * :* *
Mb2025|M.bovis_AF2122/97 DESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
Rv2002|M.tuberculosis_H37Rv DESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
Mvan_3000|M.vanbaalenii_PYR-1 DESRYATGAEFVMDGGLTNDVPHK---------------
Mflv_3281|M.gilvum_PYR-GCK DESRYSTGAEFVMDGGLTNDVPHK---------------
MSMEG_3515|M.smegmatis_MC2_155 DESRYATGAEFVMDGGLVNDVPHKL--------------
MMAR_0387|M.marinum_M DDASYATGSEFVMDGGLVTDVPHKQF-------------
MUL_1037|M.ulcerans_Agy99 DDASYATGSEFVMDGGLVTDVPHKQF-------------
MAV_5136|M.avium_104 DESSYATGSEFVMDGGLVTDVPHKQF-------------
MAB_4680c|M.abscessus_ATCC_199 DAASYVTGAELAVDGGWTAGPTLTTITGQ----------
TH_4254|M.thermoresistible__bu DDASFITGSEVYADGGWTAR-------------------
MLBr_01807|M.leprae_Br4923 EDAGYISGAVIPVDGGLGMGH------------------
: : : :*: . ***