For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDLGQRLKDKVAVITGGASGIGLATAKRMQAEGAIVVIGDVDASTGKSVANDMNVTFVQVDVADQVAVDH LFDTAFEVHGGVDIAFNNAGISPPEDDLIENTGVDAWDRVQDINLKSVFFCCKAALRHMVPAQKGSIINT ASFVAVMGSATSQISYTASKGGVLAMSRELGVQYARQGIRVNALCPGPVNTPLLQELFAKDPERAARRLV HVPVGRFAEPEELAAAVAFLASDDASFITASSFLVDGGISGHYVTPL
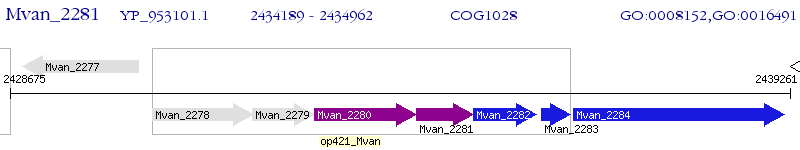
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2281 | - | - | 100% (257) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0169 | - | 2e-35 | 37.07% (259) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_1861 | - | 3e-34 | 35.36% (263) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 1e-111 | 77.43% (257) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4062 | - | 1e-137 | 93.77% (257) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv2857c | - | 1e-111 | 77.43% (257) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 3e-30 | 35.68% (241) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_1219 | - | 3e-29 | 32.39% (247) | short chain dehydrogenase |
| M. marinum M | MMAR_2981 | fabG3_1 | 2e-36 | 36.73% (245) | 20-beta-hydroxysteroid dehydrogenase FabG3_1 |
| M. avium 104 | MAV_3714 | - | 1e-110 | 76.26% (257) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2598 | - | 1e-117 | 82.49% (257) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4254 | - | 9e-37 | 35.08% (248) | PUTATIVE putative 2,5-dichloro-2,5-cyclohexadiene-1,4-diol |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 2e-34 | 36.18% (246) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2882c|M.bovis_AF2122/97 -----------MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVV
Rv2857c|M.tuberculosis_H37Rv -----------MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVV
MAV_3714|M.avium_104 -----------MMDLTQRLAGRVAVITGAGGGIGLAAARRMHAEGATIVV
Mvan_2281|M.vanbaalenii_PYR-1 ------------MDLGQRLKDKVAVITGGASGIGLATAKRMQAEGAIVVI
Mflv_4062|M.gilvum_PYR-GCK ------------MDLSQRLKDKVAVITGGASGIGLATAKRMRSEGAIVVI
MSMEG_2598|M.smegmatis_MC2_155 ------------MDLTQRLAGKVAVITGGASGIGLATGRRLRAEGATVVV
MMAR_2981|M.marinum_M ------------MAG--RLTGKVALVSGGARGMGASHVRALVAEGAHVVL
MUL_1037|M.ulcerans_Agy99 -------------MG--RVDGKVALISGGARGMGASHARLLVQEGAKVVI
TH_4254|M.thermoresistible__bu -----------MTAGSTRLAGKVVLVTGAGGGLGTAIARRLADEGARLVL
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
MAB_1219|M.abscessus_ATCC_1997 ---------------MGEFDNKVAIVTGAAQGIGEAYAHALAREGAAVVV
. .: :::*. *:* : : : :* :.:
Mb2882c|M.bovis_AF2122/97 GDVDVEAGGAAADELS--GLFVP---TDVCDEDAVNGLFDGAAETYGRID
Rv2857c|M.tuberculosis_H37Rv GDVDVEAGGAAADELS--GLFVP---TDVCDEDAVNGLFDGAAETYGRID
MAV_3714|M.avium_104 ADIDADAGAAAADELS--GLFVP---TDVADEDAVNALFDTAVRRYGRID
Mvan_2281|M.vanbaalenii_PYR-1 GDVDASTGKSVANDMN--VTFVQ---VDVADQVAVDHLFDTAFEVHGGVD
Mflv_4062|M.gilvum_PYR-GCK GDLDEKTGKSVANDLN--VTFVQ---VDVSDQVAVDNLFDTAFEVHGGVD
MSMEG_2598|M.smegmatis_MC2_155 GDIDPTTGKAAADELE--GLFVP---VDVSEQEAVDNLFDTAASTFGRVD
MMAR_2981|M.marinum_M GDILDDEGRAVAAELGDAARYVH---LDVTQPEQWTAAVDTAVNEFGGLH
MUL_1037|M.ulcerans_Agy99 GDILDEEGKALAEEIGDAARYVH---LDVTQPDQWEAAVATAVDEFGKLD
TH_4254|M.thermoresistible__bu ADLQESQCAALADELPAPAGAHHPLGFDVADETAWISAISTVSAQYGALH
MLBr_01807|M.leprae_Br4923 THRASVVPDWFFGVEC-----------DVTDNDAIGAAFKAVEEHQGPVE
MAB_1219|M.abscessus_ATCC_1997 ADINAEMGEGVAKQIVADGGRAIFAGVDVSDPDSAIAMADKAVSEFGGID
. ** : . * :.
Mb2882c|M.bovis_AF2122/97 IAFNNAGISP-PEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLA
Rv2857c|M.tuberculosis_H37Rv IAFNNAGISP-PEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLA
MAV_3714|M.avium_104 IAFNNAGISP-PDDDVIENTEPPAWQRVQDVNLKSVYLCCRAALRHMVSA
Mvan_2281|M.vanbaalenii_PYR-1 IAFNNAGISP-PEDDLIENTGVDAWDRVQDINLKSVFFCCKAALRHMVPA
Mflv_4062|M.gilvum_PYR-GCK IAFNNAGISP-PEDDLIENTGIDAWDRVQDINLKSVFFCCKAALRHMVPA
MSMEG_2598|M.smegmatis_MC2_155 IAFNNAGISP-PEDDLIENTDLPAWQRVQDINLKSVYLSCRAALRHMVPA
MMAR_2981|M.marinum_M VLVNNAGILN-IGT--IEDYALSEWQRILDINVTGVFLGIRAAVKPMKEA
MUL_1037|M.ulcerans_Agy99 VLVNNVGIVA-LGQ--LKKFDLGKWQKVIDVNLTGTFLGMRAAVEPMTAA
TH_4254|M.thermoresistible__bu ALVNNAAIGA-LGN--VAEERLERWDRILDVGATGVWLGMKHAVPLIRRS
MLBr_01807|M.leprae_Br4923 VLVANAGIT---SDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQ
MAB_1219|M.abscessus_ATCC_1997 YLVNNAAIYGGMKLDLLLSVPWDYYKKFMSVNQDGALVCTRAVYKHIAQR
. *..* : :.:. .:. .. : . :
Mb2882c|M.bovis_AF2122/97 GKGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVN
Rv2857c|M.tuberculosis_H37Rv GKGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVN
MAV_3714|M.avium_104 RRGSIINTASFVAVMGSATSQISYTAAKGGVLALSRELGVQFARQGIRVN
Mvan_2281|M.vanbaalenii_PYR-1 QKGSIINTASFVAVMGSATSQISYTASKGGVLAMSRELGVQYARQGIRVN
Mflv_4062|M.gilvum_PYR-GCK QKGSIVNTASFVAVNGSATSQISYTASKGGVLAMSRELGIQYARQGIRVN
MSMEG_2598|M.smegmatis_MC2_155 GKGSIINTASFVAVMGSATSQISYTASKGGVLAMSRELGVQYARQGIRVN
MMAR_2981|M.marinum_M GRGSIINISSIEG-LAGTIASHGYTVSKFAVRGLTKSTALELGPSGIRVN
MUL_1037|M.ulcerans_Agy99 GSGSIINVSSIEG-LRGAPAVHPYVASKWAVRGLTKSAALELAPLNIRVN
TH_4254|M.thermoresistible__bu GGGSVVNICSIFGTVGGFGDSAAYHAAKGAVRTLTKNAALHWAPDGVRVN
MLBr_01807|M.leprae_Br4923 RFGRYIFMGSVVG-LSGVPGQANYAASKSGLIGMARSLARELGSRSITAN
MAB_1219|M.abscessus_ATCC_1997 GGGAIVNQSSTAAWLYSG----FYGLAKVGINGLTQQLSRELGGQKIRIN
* : * . . * :* .: :::. . . . : *
Mb2882c|M.bovis_AF2122/97 ALCPGPVNTPLLQELFAKNPERAARRMVHVPLGRFAEPDEIAAAVAFLAS
Rv2857c|M.tuberculosis_H37Rv ALCPGPVNTPLLQELFAKNPERAARRMVHVPLGRFAEPDEIAAAVAFLAS
MAV_3714|M.avium_104 ALCPGPVNTPLLQELFAKDPQRAARRLVHVPVGRFAEPGEIAAAAAFLAS
Mvan_2281|M.vanbaalenii_PYR-1 ALCPGPVNTPLLQELFAKDPERAARRLVHVPVGRFAEPEELAAAVAFLAS
Mflv_4062|M.gilvum_PYR-GCK ALCPGPVNTPLLQELFAKDPERAARRLVHVPMGRFAEPEELAAAVAFLAS
MSMEG_2598|M.smegmatis_MC2_155 ALCPGPVNTPLLQELFAKDPERAARRLVHIPLGRFAEPEELAAAVAFLAS
MMAR_2981|M.marinum_M SIHPGLVKTP--------MTEWVPEDLFQTALGRAAEPMEVSNLVVYLAS
MUL_1037|M.ulcerans_Agy99 SIHPGFIRTP--------MTANLPDDMVTIPLGRPAESREVSTFVVFLAS
TH_4254|M.thermoresistible__bu SLHPGFIETPQLRELYK-GTERYERMLAGTPLGRLGRPDEIAAVVAFLVS
MLBr_01807|M.leprae_Br4923 VVAPGFIDTEMTR---AMDSKYQEAALQAIPLGRIGAVEEIAGAVSFLAS
MAB_1219|M.abscessus_ATCC_1997 AIAPGPTDTEATRG--TVPEQFRSELVKNIPLSRMGTVDDMVGMCLFLLS
: ** * : .:.* . :: :* *
Mb2882c|M.bovis_AF2122/97 DDASFITASTFLVDGGISSAYVTPL--------------
Rv2857c|M.tuberculosis_H37Rv DDASFITASTFLVDGGISSAYVTPL--------------
MAV_3714|M.avium_104 DDASFITASTFLVDGGISAAYVTPL--------------
Mvan_2281|M.vanbaalenii_PYR-1 DDASFITASSFLVDGGISGHYVTPL--------------
Mflv_4062|M.gilvum_PYR-GCK DDASFITGSSFLVDGGITGHYVTPL--------------
MSMEG_2598|M.smegmatis_MC2_155 DDASFITGSTFLVDGGISSAYVTPL--------------
MMAR_2981|M.marinum_M DESSYSTGAEFVVDGGTVAGLAHKDFSAVDVAAQPEWVT
MUL_1037|M.ulcerans_Agy99 DDASYATGSEFVMDGGLVTDVPHKQF-------------
TH_4254|M.thermoresistible__bu DDASFITGSEVYADGGWTAR-------------------
MLBr_01807|M.leprae_Br4923 EDAGYISGAVIPVDGGLGMGH------------------
MAB_1219|M.abscessus_ATCC_1997 DKASWITGQVFNVDGGQIIRS------------------
:.:.: :. . ***