For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKIDLTGKTALVTGSTQGIGLAIARSLAASGARVAVNGRSRDRVDEAVATLDGDVVGVAADVADAAGAQA LAAELPHIDILVNNLGIFGAVPAREITDDQWRSYFEVNVLAAARLIRHYLPGMVDRGWGRVLQIASDSAI VIPEEMIHYGVSKTALLALSRGFAKDASGTGVTVNSVIAGPTHTAGVEDFVYQLVDKSLPWEQAQREFMR LHRPQSLIERLIEPEEIANMVTYLASPLASATTGGALRVDGGYVDAILP
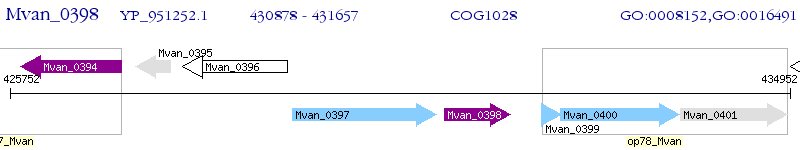
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0398 | - | - | 100% (259) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_5918 | - | 1e-28 | 33.71% (264) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1212 | - | 9e-27 | 34.80% (250) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1963c | - | 2e-25 | 31.15% (260) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0296 | - | 8e-43 | 37.84% (259) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1928c | - | 2e-25 | 31.15% (260) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 2e-21 | 31.23% (253) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_4665c | - | 1e-102 | 75.21% (238) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0629 | - | 7e-27 | 33.85% (257) | dehydrogenase |
| M. avium 104 | MAV_2438 | - | 4e-30 | 34.22% (263) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_0600 | - | 1e-122 | 84.23% (260) | dehydrogenase |
| M. thermoresistible (build 8) | TH_3893 | - | 1e-34 | 34.72% (265) | PUTATIVE putative short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0315 | fabG2 | 2e-22 | 31.45% (248) | 3-oxoacyl- |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0315|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0629|M.marinum_M --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0398|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0600|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4665c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_0296|M.gilvum_PYR-GCK --------------------------------------------------
TH_3893|M.thermoresistible__bu --------------------------------------------------
MAV_2438|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
MUL_0315|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0629|M.marinum_M --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0398|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0600|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4665c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_0296|M.gilvum_PYR-GCK --------------------------------------------------
TH_3893|M.thermoresistible__bu --------------------------------------------------
MAV_2438|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
MUL_0315|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0629|M.marinum_M --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0398|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0600|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4665c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_0296|M.gilvum_PYR-GCK --------------------------------------------------
TH_3893|M.thermoresistible__bu --------------------------------------------------
MAV_2438|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
MUL_0315|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0629|M.marinum_M --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0398|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0600|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4665c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_0296|M.gilvum_PYR-GCK --------------------------------------------------
TH_3893|M.thermoresistible__bu --------------------------------------------------
MAV_2438|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
MUL_0315|M.ulcerans_Agy99 --MRPDSSPSWCTITSVVTGGASGIGLAIARRLSADGTDVATLDLKTPDG
MMAR_0629|M.marinum_M ----------MAGLTALITGATSGIGRATAHGLAELGATVLVSGRDEARG
Mb1963c|M.bovis_AF2122/97 --MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQVAIAARHLDAL
Rv1928c|M.tuberculosis_H37Rv --MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQVAIAARHLDAL
Mvan_0398|M.vanbaalenii_PYR-1 ------MKIDLTGKTALVTGSTQGIGLAIARSLAASGARVAVNGRSRDRV
MSMEG_0600|M.smegmatis_MC2_155 ------MNIDLTGKTALVTGSTQGIGLAIAETLARSGARVAINGRTASRV
MAB_4665c|M.abscessus_ATCC_199 -----------------------------MRQLAASGARAVVNGRSQERV
Mflv_0296|M.gilvum_PYR-GCK ------MDMGLSGKRALVTGSSSGLGRAIAESLAAEGASVVVHGRDEVRT
TH_3893|M.thermoresistible__bu ------MDFDLNGKRALVTGSSSGIGAGIVETLAAEGVSVVVHGRNAERA
MAV_2438|M.avium_104 ------MDLGLAGSTAVVTGGSKGMGLAVAETLAAEGAKVAVMARGRHAL
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVESAAE
*. .
MUL_0315|M.ulcerans_Agy99 --------------GLAFSADVTNRSQVD-AALAAIRQRLGPVTILVNAA
MMAR_0629|M.marinum_M REVVDEVTAR-GGRGIFLAAELRDVTGVRNLATAAADAGAGKVDILINSA
Mb1963c|M.bovis_AF2122/97 EKLADEIGTS-GGKVVPVCCDVSQHQQVT-SMLDQVTAELGGIDIAVCNA
Rv1928c|M.tuberculosis_H37Rv EKLADEIGTS-GGKVVPVCCDVSQHQQVT-SMLDQVTAELGGIDIAVCNA
Mvan_0398|M.vanbaalenii_PYR-1 DEAVATL----DGDVVGVAADVADAAGAQ-ALAAELP----HIDILVNNL
MSMEG_0600|M.smegmatis_MC2_155 DETVAQLG---DLDVVGVAADVATQKGTE-ALLEQLP----SVDILVNNL
MAB_4665c|M.abscessus_ATCC_199 DAAVAEVGLQ-HGSVSGVAADVSTADGVE-RLLKQLP----EVDILINNL
Mflv_0296|M.gilvum_PYR-GCK GAVADGIRAR-GGDAWAAVGDLATEGGAG-AVAAAAG----EVDILVNNA
TH_3893|M.thermoresistible__bu EAVADRIRAD-GGTAATAVGDLATEDGAA-QVADTAAAAFSGIDILVNNA
MAV_2438|M.avium_104 DAAVALLREAGAPDAVGVSVDMADAESIA-DGFAAVAQRWGRLNSLVHTI
MLBr_02565|M.leprae_Br4923 ALDETANRVG----GTTLSLDVTADDAVDKITEHLHDYQGGHADILVNNA
:: :
MUL_0315|M.ulcerans_Agy99 GLDGFKR---FSNITFEDWQRVIDVNLNGVFHTIQAVLPDMVAAGWG-RI
MMAR_0629|M.marinum_M GAFPFGP---TADTSPDDFDAVFALNVRAPYFLVGALAPTMAKRGRG-SI
Mb1963c|M.bovis_AF2122/97 GIITVTP---MLDMPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQGQGGVI
Rv1928c|M.tuberculosis_H37Rv GIITVTP---MLDMPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQGQGGVI
Mvan_0398|M.vanbaalenii_PYR-1 GIFGAVP---AREITDDQWRSYFEVNVLAAARLIRHYLPGMVDRGWG-RV
MSMEG_0600|M.smegmatis_MC2_155 GIFGAEP---AREITDEQWRKYFDVNVLAAVRLIRSYLPGMIEKGWG-RA
MAB_4665c|M.abscessus_ATCC_199 GIFGATP---AREITDEQWRNYFEVNVLSAVRLIRAYLPAMIGAGWG-RI
Mflv_0296|M.gilvum_PYR-GCK GVYDG---LAWSDLAPRQWQSIYEVNVVSGVRMIEYLVPGMRQRGWG-RI
TH_3893|M.thermoresistible__bu GGSSETGIQSWFALPVSEWGVTYQRNVMSAGYLIHALVPAMKESGWG-RI
MAV_2438|M.avium_104 GPGDGD----FEELDDTGWDEAFALGTMSAVRSIRNALPLLRAADWG-RI
MLBr_02565|M.leprae_Br4923 GITRDKL---LANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSISKGG-RV
* : . . *
MUL_0315|M.ulcerans_Agy99 VNISSSSTHSG--TPYMTHYVAAKSAVNGLTKALALEYGPSGITVNAVPP
MMAR_0629|M.marinum_M VNVTTMVAEFG--AAGTGLYGASKAAIALLTKSWAAEYGPSGVRVNAVSP
Mb1963c|M.bovis_AF2122/97 INTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPHKIRVNSVSP
Rv1928c|M.tuberculosis_H37Rv INTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPHKIRVNSVSP
Mvan_0398|M.vanbaalenii_PYR-1 LQIASDSAIVI--PEEMIHYGVSKTALLALSRGFAKDASGTGVTVNSVIA
MSMEG_0600|M.smegmatis_MC2_155 IQIASDSAIVI--PEEMIHYGVSKTALLAVSRGFAKDAAGTGVTVNSVIA
MAB_4665c|M.abscessus_ATCC_199 IQIASDSAIVI--PQEMIHYGVSKTALLGVTRGFAKDAAGSGVTINSVIA
Mflv_0296|M.gilvum_PYR-GCK IQIGGGLALQP--AADQPHYNATLAARHNLTVSLARELAGTGVTSNTVAP
TH_3893|M.thermoresistible__bu IQIASAAGIIP--TSGQPDYGPSKAAMINMSMGLSKALAGTGITVNTVMP
MAV_2438|M.avium_104 VTFAAHSIQRQ--NPRIVAYTASKAALASITKNLSKSLAAEGILVNCVCP
MLBr_02565|M.leprae_Br4923 IVLSSMAGIAG--NRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAP
: * : :. :: : . : * * .
MUL_0315|M.ulcerans_Agy99 GFIDTPMLRNAAAQGHLG--------------DIEQNIARTPVRRIGKPE
MMAR_0629|M.marinum_M GPTRTEGTVEMGEA-------------------LDQLAAAAPAGRPADPT
Mb1963c|M.bovis_AF2122/97 GYILTELVEPYTEY-------------------QPLWEPKIPLGRLGRPE
Rv1928c|M.tuberculosis_H37Rv GYILTELVEPYTEY-------------------QPLWEPKIPLGRLGRPE
Mvan_0398|M.vanbaalenii_PYR-1 GPTHTAGVEDFVYQLVDK---SLPWEQAQREFMRLHRPQS-LIERLIEPE
MSMEG_0600|M.smegmatis_MC2_155 GPTHTAGVEDFVYQLVDK---SLPWDEAQREFMRKHRPQS-LLQRLIEPE
MAB_4665c|M.abscessus_ATCC_199 GPTHTAGVEDFVYQLVDK---SLPWDEAQREFMKRHRPQS-LVQRLIEPA
Mflv_0296|M.gilvum_PYR-GCK GAILTEPVREMTYRVAAQRGWGPTWEDVEKAAATTWFPN--DIGRFGRPE
TH_3893|M.thermoresistible__bu GMIMTEGLRDFLRTFADHRGWG---DDLDKAADYILKGTGQTVHRIGQVS
MAV_2438|M.avium_104 GTIVTASFTEALKGVLAADGLDATDPVDVMTWIDDNFHQPCDLGRAGLPE
MLBr_02565|M.leprae_Br4923 GFIETQMTAAIPLAT------------------REVGRRLNSLLQGGLPV
* * :
MUL_0315|M.ulcerans_Agy99 DIAAACAFLVSEEAGYITGQILGVNGGRNT----
MMAR_0629|M.marinum_M EIASTIVYLASDAASFIHGAVVPVDGGRAAV---
Mb1963c|M.bovis_AF2122/97 ELAGLYLYLASEASSYMTGSDIVIDGGYTCP---
Rv1928c|M.tuberculosis_H37Rv ELAGLYLYLASEASSYMTGSDIVIDGGYTCP---
Mvan_0398|M.vanbaalenii_PYR-1 EIANMVTYLASPLASATTGGALRVDGGYVDAILP
MSMEG_0600|M.smegmatis_MC2_155 EIANMVAYLASPLASATTGGALRVDGGYVDAILP
MAB_4665c|M.abscessus_ATCC_199 EIANLVVYLSSPLASATTGAAVRADGGYVDSILP
Mflv_0296|M.gilvum_PYR-GCK EVAAAVAFLAGVHAGYISGADLRVDGGAIRSVA-
TH_3893|M.thermoresistible__bu DVAYAVAFLASPRADFLNGINLHLDGGATQSIY-
MAV_2438|M.avium_104 EVASITVYLASRRNGYVTGAMVNVDGGSDFI---
MLBr_02565|M.leprae_Br4923 DVAETIAYFAIPASNAVTGNVIRVCGQAMLGA--
::* :: . * : *