For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQVAIAARHLDALEKLADEIGTSGGKVVPVCCDVS QHQQVTSMLDQVTAELGGIDIAVCNAGIITVTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQGQG GVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPHKIRVNSVSPGYILTELVEPYTEYQPL WEPKIPLGRLGRPEELAGLYLYLASEASSYMTGSDIVIDGGYTCP
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
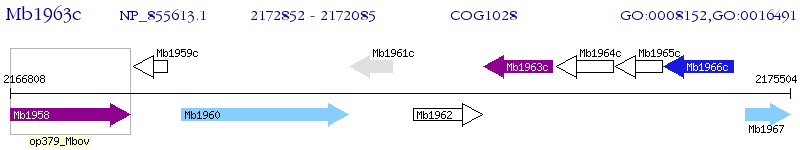
| M. bovis AF2122 / 97 | Mb1963c | - | - | 100% (255) | short chain dehydrogenase |
| M. bovis AF2122 / 97 | Mb0950c | - | 2e-35 | 33.98% (256) | short chain dehydrogenase |
| M. bovis AF2122 / 97 | Mb1385 | fabG | 5e-35 | 34.96% (246) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0305 | - | 1e-40 | 38.10% (252) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1928c | - | 1e-144 | 100.00% (255) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 2e-30 | 35.10% (245) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4490c | - | 8e-40 | 36.29% (259) | oxidoreductase |
| M. marinum M | MMAR_2849 | - | 1e-118 | 81.96% (255) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_2775 | - | 1e-108 | 75.98% (254) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0779 | - | 2e-44 | 38.98% (254) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_2623 | - | 3e-43 | 38.82% (255) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3898 | fabG | 2e-36 | 36.51% (241) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_0441 | - | 9e-43 | 38.00% (250) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0305|M.gilvum_PYR-GCK ------MTTTS--ANPFDLTGHVAVVTGGGSGIGLGMAGGLARAGATVAI
Mvan_0441|M.vanbaalenii_PYR-1 ------MTDAAPPANPFDLTGHVAVVTGGGSGIGLGMAEGLARAGATVAI
Mb1963c|M.bovis_AF2122/97 ----------MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQVAI
Rv1928c|M.tuberculosis_H37Rv ----------MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQVAI
MMAR_2849|M.marinum_M ----------MSVLDLFDLSGKRALITGASTGIGKKVALAYAEAGAQVAV
MAV_2775|M.avium_104 ------------MLDLFDLRGRRALVTGASTGIGKQVAQAYLQAGARVAI
MAB_4490c|M.abscessus_ATCC_199 --------MFDDPRALFDIAGKSAIVIGATGSLGQIASRALAGAGANVAI
TH_2623|M.thermoresistible__bu ----------MGVLDKFRLDDKVVIVTGASSGLGVAFARACAEAGADVVL
MSMEG_0779|M.smegmatis_MC2_155 ---------------MFDLQGRSVVVTGGTKGIGRGIATVFARAGANVAV
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
MUL_3898|M.ulcerans_Agy99 ------MCGAHRDRG-----YVYPVVTGGAQGLGLAIAERFVSEGARVVL
:: *. .:* : * *.:
Mflv_0305|M.gilvum_PYR-GCK LGRNAQRLDDAAATLRTHG-NPVLPLVCDVTDEEAVTETMARIRDEFGWL
Mvan_0441|M.vanbaalenii_PYR-1 LGRSAQRLDSAAATLRSHG-NPVLPVVCDVTDEQAVTDAMARIRSEFGWL
Mb1963c|M.bovis_AF2122/97 AARHLDALEKLADEIGTSG-GKVVPVCCDVSQHQQVTSMLDQVTAELGGI
Rv1928c|M.tuberculosis_H37Rv AARHLDALEKLADEIGTSG-GKVVPVCCDVSQHQQVTSMLDQVTAELGGI
MMAR_2849|M.marinum_M AARHSDALQVVADEIAGVG-GKALPIRCDVTQPDQVRGMLDQMTGELGGI
MAV_2775|M.avium_104 AARDFEALQRTADELTAGGTGGVVAIRCDVTQPDEVSGMVDRMIAELGGI
MAB_4490c|M.abscessus_ATCC_199 CGSREDALSDLRDELAAAP-GCIVPIARRPDSESASDAIVKETIEAFGGV
TH_2623|M.thermoresistible__bu AARRVERLQQTAEQVRATG-RRALTVATDVVDPQQCQEMVDAAMTEFGRV
MSMEG_0779|M.smegmatis_MC2_155 AGRSTADIDACVADLDQLGSGKVIGVQTDVSDRAQCDALAGRAVEEFGGI
MLBr_01807|M.leprae_Br4923 THR---ASVVPDWFFG---------VECDVTDNDAIGAAFKAVEEHQGPV
MUL_3898|M.ulcerans_Agy99 GDVNLEATEAAAKQLGGGE--VAVAVRCDVTKADEVETLLQAALERFGGL
. : . * :
Mflv_0305|M.gilvum_PYR-GCK DSCFANAGVRGRFTPVLETSLAEFRSVTSVDLDGVFLTLREAARQMVAAG
Mvan_0441|M.vanbaalenii_PYR-1 DSCFANAGVRGTFTPTLDTSLAEFREVTRVDLDGVFVTLREAARQMIGAG
Mb1963c|M.bovis_AF2122/97 DIAVCNAGII-TVTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQG
Rv1928c|M.tuberculosis_H37Rv DIAVCNAGII-TVTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQG
MMAR_2849|M.marinum_M DIAVCNAGIV-SVQAMLDMPLEEFQRIQDTNVTGVFLTAQAAARAMVDQG
MAV_2775|M.avium_104 DIAVCNAGII-SVSPMLEMPAAEFQRIQDTNVTGVFLTAQAAARAMVRQG
MAB_4490c|M.abscessus_ATCC_199 DILVVASGVN-EVAPAVDMQPARFEAVLDANVTSTWLAVRSAGRAMIERG
TH_2623|M.thermoresistible__bu DVLVNNAGVG-TAVPATRETPEEFRTVIDINLNGSYWAAQACGRVMRP--
MSMEG_0779|M.smegmatis_MC2_155 DVVCANAGVF-PDAPLATMTPEQLNGIFAVNVNGTFYAVQACLDALIASG
MLBr_01807|M.leprae_Br4923 EVLVANAGIT-SDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR
MUL_3898|M.ulcerans_Agy99 DIMVNNAGIT-RDATMRKMTEEQFDQVIDVHLKGTWNGIRLAAAIMRENK
: :*: .: : .: . : : . :
Mflv_0305|M.gilvum_PYR-GCK RGGSLVGVSSLGAFQG--MPRQPAYAASKAGVTSMMDSFAVELAR-HGIR
Mvan_0441|M.vanbaalenii_PYR-1 RGGSLVGVSSLGAVQG--MPRQPAYAASKAGVTSLMDSLAVEFAR-YGIR
Mb1963c|M.bovis_AF2122/97 QGGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAP-HKIR
Rv1928c|M.tuberculosis_H37Rv QGGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAP-HKIR
MMAR_2849|M.marinum_M LGGTIITTASMSGHIINIPQQVSHYCTSKAAVVHLTKAMAVELAP-HQIR
MAV_2775|M.avium_104 RGGAIITTASMSGHIINVPQQVGHYCASKAAVIQLTKAMAVEFAP-HDIR
MAB_4490c|M.abscessus_ATCC_199 RGGKIILTSSARGKLG-HPAGYTAYCASKAAVDGLVRSFGCEWGR-YGIT
TH_2623|M.thermoresistible__bu -GSSIVNISSILGITT-ASLPQAAYSASKAGVIGLTRDLAQQWTPRKGIR
MSMEG_0779|M.smegmatis_MC2_155 -SGRVVLTSSITGPITGYPGWS-HYGATKAAQLGFMRTAAIELAP-HKIT
MLBr_01807|M.leprae_Br4923 -FGRYIFMGSVVGLSG--VPGQANYAASKSGLIGMARSLARELGS-RSIT
MUL_3898|M.ulcerans_Agy99 -RGAIVNMSSVSGKVG--MVGQTNYSAAKAGIVGMTKAAAKELAH-VGVR
. : .* . * ::*:. : . : :
Mflv_0305|M.gilvum_PYR-GCK ANTVAPGWFETEMT----SEGMADERFTSRVLPRVPVRRWGAAEDVAGVG
Mvan_0441|M.vanbaalenii_PYR-1 ANTIQPGWFATEMT----ADGMADERFRARVLPRVPVRRWGAAEDVG--G
Mb1963c|M.bovis_AF2122/97 VNSVSPGYILTELV------EPYTEYQPL-WEPKIPLGRLGRPEELA--G
Rv1928c|M.tuberculosis_H37Rv VNSVSPGYILTELV------EPYTEYQPL-WEPKIPLGRLGRPEELA--G
MMAR_2849|M.marinum_M VNSVSPGYIRTELV------EPLADYHAL-WEPKIPLGRMGRPEELT--G
MAV_2775|M.avium_104 VNSVSPGYIRTELV------EPLHEYHRQ-WQPKIPLGRMGRPDELA--G
MAB_4490c|M.abscessus_ATCC_199 VNGLAPTVFRSPLTSWMFTDEPAAQATRENFLSRIPLGRLGEPEDLA--G
TH_2623|M.thermoresistible__bu VNAIAPGFFKSEMT------DQYKPGYLDSQMPRVLMGRTGDPEELA--A
MSMEG_0779|M.smegmatis_MC2_155 VNAIMPGNIMTEGL------LENGEEYIASMARSIPAGALGTPEDIG--H
MLBr_01807|M.leprae_Br4923 ANVVAPGFIDTEMT------RAMDSKYQEAALQAIPLGRIGAVEEIA--G
MUL_3898|M.ulcerans_Agy99 VNAIAPGLIRSAMT------EAMPQRIWDQKLAEVPMGRAGEPSEVA--S
.* : * : : : * .::
Mflv_0305|M.gilvum_PYR-GCK VAVYLAGPASAYHTGDVLRLDGGYLKF--------
Mvan_0441|M.vanbaalenii_PYR-1 IAVYLASPASAYHTGDVLRLDGGYLKF--------
Mb1963c|M.bovis_AF2122/97 LYLYLASEASSYMTGSDIVIDGGYTCP--------
Rv1928c|M.tuberculosis_H37Rv LYLYLASEASSYMTGSDIVIDGGYTCP--------
MMAR_2849|M.marinum_M LYLYLASAASSYMTGSDIVIDGGYTCP--------
MAV_2775|M.avium_104 IYLYLASAASSYMTGSDIVIDGGYSCP--------
MAB_4490c|M.abscessus_ATCC_199 PILFLSAPASDFLTGHVVYPDGGYTAG--------
TH_2623|M.thermoresistible__bu TLVWLVSDAGGYVTGQTIVVDGGITIT--------
MSMEG_0779|M.smegmatis_MC2_155 LAAFLATKEAGYITGQAIAVDGGQVLPESLDAIAT
MLBr_01807|M.leprae_Br4923 AVSFLASEDAGYISGAVIPVDGGLGMGH-------
MUL_3898|M.ulcerans_Agy99 VALFLACDLSSYMTGTVLDVTGGRFI---------
:* . : :* : **