For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DLGLAGSTAVVTGGSKGMGLAVAETLAAEGAKVAVMARGRHALDAAVALLREAGAPDAVGVSVDMADAES IADGFAAVAQRWGRLNSLVHTIGPGDGDFEELDDTGWDEAFALGTMSAVRSIRNALPLLRAADWGRIVTF AAHSIQRQNPRIVAYTASKAALASITKNLSKSLAAEGILVNCVCPGTIVTASFTEALKGVLAADGLDATD PVDVMTWIDDNFHQPCDLGRAGLPEEVASITVYLASRRNGYVTGAMVNVDGGSDFI*
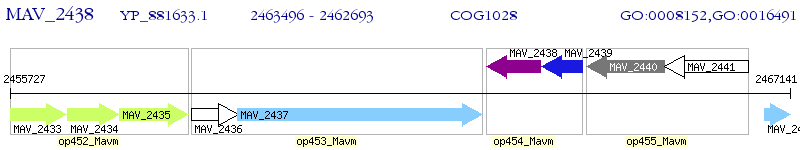
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2438 | - | - | 100% (267) | oxidoreductase, short chain dehydrogenase/reductase family protein |
| M. avium 104 | MAV_1587 | - | 6e-70 | 51.87% (268) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. avium 104 | MAV_1891 | - | 3e-34 | 34.72% (265) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1385 | fabG | 8e-25 | 30.27% (261) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_4353 | - | 2e-70 | 52.99% (268) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1350 | fabG | 8e-25 | 30.27% (261) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-18 | 28.14% (263) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0833 | - | 1e-26 | 31.46% (267) | Short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_1568 | - | 1e-122 | 80.52% (267) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_2228 | - | 3e-73 | 51.87% (268) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_4608 | - | 4e-68 | 50.93% (269) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0315 | fabG2 | 4e-25 | 30.74% (257) | 3-oxoacyl- |
| M. vanbaalenii PYR-1 | Mvan_1991 | - | 5e-69 | 51.87% (268) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1385|M.bovis_AF2122/97 --------------MASLLNARTAVITGGAQGLGLAIGQRFVAEGARVVL
Rv1350|M.tuberculosis_H37Rv --------------MASLLNARTAVITGGAQGLGLAIGQRFVAEGARVVL
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
MUL_0315|M.ulcerans_Agy99 ---------MRPD-SSPSWCTITSVVTGGASGIGLAIARRLSADGTDVAT
MAB_0833|M.abscessus_ATCC_1997 -------MNRETFSRLFDLTGRTAIVTGGTRGIGRALAEGYACAGANVVV
MAV_2438|M.avium_104 --------------MDLGLAGSTAVVTGGSKGMGLAVAETLAAEGAKVAV
MMAR_1568|M.marinum_M --------------MDLGFSGSKAVVTGGSKGMGLAIAETLAAEGASVAV
Mflv_4353|M.gilvum_PYR-GCK --------------MDLGLRDATVVVVGGGRGMGLAAAHCFADDGARVAL
Mvan_1991|M.vanbaalenii_PYR-1 --------------MDLGLRDATAVVVGGGRGMGLAAAHCFADDGARVAL
TH_4608|M.thermoresistible__bu --------------MDLGFTDAAAVVVGGGRGMGLAAAQCLADDGARVAV
MSMEG_2228|M.smegmatis_MC2_155 --------------MDLGFGGAAAAVVGGGRGMGFATAQCLADDGARVAV
:.** *:* * .. * *.
Mb1385|M.bovis_AF2122/97 GDVNL-EATEVAAKRL-GGDDVALAVRCDVTQADDVDILIRTAVERFG-G
Rv1350|M.tuberculosis_H37Rv GDVNL-EATEVAAKRL-GGDDVALAVRCDVTQADDVDILIRTAVERFG-G
MLBr_01807|M.leprae_Br4923 -----------THRAS-VVPDWFFGVECDVTDNDAIGAAFKAVEEHQG-P
MUL_0315|M.ulcerans_Agy99 -----------LDLK---TPDGGLAFSADVTNRSQVDAALAAIRQRLG-P
MAB_0833|M.abscessus_ATCC_1997 ASRKPEACTEAVERLR-SLGARALGVPTHTGDIHSLEELVRATAEEFG-G
MAV_2438|M.avium_104 MARGRHALDAAVALLREAGAPDAVGVSVDMADAESIADGFAAVAQRWG-R
MMAR_1568|M.marinum_M IARGKRALDSAVERLSQAGAPDAVAISADMADAASITSAFAAVSERWG-Q
Mflv_4353|M.gilvum_PYR-GCK VGRTRDVLDEAAAQLRGRGSPDAVGVVADCADEAAVQRAFGELAERWNGE
Mvan_1991|M.vanbaalenii_PYR-1 IGRSPEVLDGAAAQLRERGSPDAVGVVADTTDDAAVRRAFAELGERWNGE
TH_4608|M.thermoresistible__bu IGRTRSMLDEAAADLARRGSPDALGIVADIRDEQQVERVFAELGERWQGE
MSMEG_2228|M.smegmatis_MC2_155 IGRSKDVLETAASDLARRGSPDAIAIVADTGDAEQVERAFAEIGERWDGE
... . : : . :.
Mb1385|M.bovis_AF2122/97 LDVMVNNAGITRDATMRTMTEEQFDQVIAVHLKGTWNGTRLAAAIMRERK
Rv1350|M.tuberculosis_H37Rv LDVMVNNAGITRDATMRTMTEEQFDQVIAVHLKGTWNGTRLAAAIMRERK
MLBr_01807|M.leprae_Br4923 VEVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR
MUL_0315|M.ulcerans_Agy99 VTILVNAAGLDGFKRFSNITFEDWQRVIDVNLNGVFHTIQAVLPDMVAAG
MAB_0833|M.abscessus_ATCC_1997 IDIVVNNAANALTEPVGAFTVDGWDKSFGVNLRGPVFLVQAALPHLVASP
MAV_2438|M.avium_104 LNSLVHTIGPGD-GDFEELDDTGWDEAFALGTMSAVRSIRNALPLLRAAD
MMAR_1568|M.marinum_M LNCLVHTIGPGD-GYFEELDDAAWDAAFTLGTMSGVRAIRAALPLLRAAD
Mflv_4353|M.gilvum_PYR-GCK LNVLVNAVGPTVRGTFDELSDADWRQAVDEGAMGMVRCVRAALPLLRAAE
Mvan_1991|M.vanbaalenii_PYR-1 LNILVNAVGPTVRGTFDELTDADWRQAVDEGAMGMVHCVRAALPLLRAAE
TH_4608|M.thermoresistible__bu LNVLINTVGPAVMGTVEELSDEQWRSAVDDGVLGMVHCVRSALPLLRQAE
MSMEG_2228|M.smegmatis_MC2_155 LNVLINTVGPGAAGDFEELTDGQWQEAFDAGVMGMVRCVRAALPLLRKAE
: :: . . : : . . : . :
Mb1385|M.bovis_AF2122/97 RGAIVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAAKELAHLGIRVNAI
Rv1350|M.tuberculosis_H37Rv RGAIVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAAKELAHLGIRVNAI
MLBr_01807|M.leprae_Br4923 FGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITANVV
MUL_0315|M.ulcerans_Agy99 WGRIVNISSSSTHSGTPYMTHYVAAKSAVNGLTKALALEYGPSGITVNAV
MAB_0833|M.abscessus_ATCC_1997 HAAVLNVVSVGAFMFGQGVSMYSAAKAALVSYTRSAAAELASRGVRVNAL
MAV_2438|M.avium_104 WGRIVTFAAHSIQRQNPRIVAYTASKAALASITKNLSKSLAAEGILVNCV
MMAR_1568|M.marinum_M WARVVTLSAHSIQRQNPRLVAYTASKAALSSVTKNLSKSLAKEGILVNCV
Mflv_4353|M.gilvum_PYR-GCK WARIVNFSAHSTQRQSVILPAYTAAKAMVTSISKNLSLLLARDEILVNVV
Mvan_1991|M.vanbaalenii_PYR-1 WARIVNFSAHSTQRQSVILPAYTAAKAMVTSISKNLSLLLARDEILVNVV
TH_4608|M.thermoresistible__bu WARIVNLSAHSTQRQSVILPAYTAAKAMVTSISKNLSLLLAKDEILVNVV
MSMEG_2228|M.smegmatis_MC2_155 WARIVNFSAHSTQRQSTRLPAYTAAKAAVNSVSKNLSLLLAKDEIMVNVV
. : . : * *:*: : . :: : . : .* :
Mb1385|M.bovis_AF2122/97 APGLIRS--------------AMTEAMPQR----IWDQKLAEVPMGRAGE
Rv1350|M.tuberculosis_H37Rv APGLIRS--------------AMTEAMPQR----IWDQKLAEVPMGRAGE
MLBr_01807|M.leprae_Br4923 APGFIDT--------------EMTRAMDSK----YQEAALQAIPLGRIGA
MUL_0315|M.ulcerans_Agy99 PPGFIDT--------------PMLRNAAAQGHLGDIEQNIARTPVRRIGK
MAB_0833|M.abscessus_ATCC_1997 APGAIDT--------------DMVRKTPPA----EVERMANTNHLKRLGL
MAV_2438|M.avium_104 CPGTIVTASFTEALKGVLAADGLDATDPVDVMTWIDDNFHQPCDLGRAGL
MMAR_1568|M.marinum_M CPGTIVTASFTEQLKDLLAADGLDATNPYDVMAWIDKYFHQPCDLGRAGL
Mflv_4353|M.gilvum_PYR-GCK SPGSIAS----EALVGWADSVGVDGSDPYALMAAIDEHFGHPAHLPRAGL
Mvan_1991|M.vanbaalenii_PYR-1 SPGSIAS----EALLGWADSVGVDGSDPYALMAAIDEHFGHPAHLPRAGL
TH_4608|M.thermoresistible__bu SPGSIAS----EALVGWADSIGVDGTDPYALMSAIDEHFGHPAHLPRAGL
MSMEG_2228|M.smegmatis_MC2_155 SPGSISS----EALVGWAESVGVDGSDPYQLMAAIDEHFGHPAHLPRAGL
** * : : . : * *
Mb1385|M.bovis_AF2122/97 PSEVASVAVFLASDLSSYMTGTVLDVTGGRFI--
Rv1350|M.tuberculosis_H37Rv PSEVASVAVFLASDLSSYMTGTVLDVTGGRFI--
MLBr_01807|M.leprae_Br4923 VEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH
MUL_0315|M.ulcerans_Agy99 PEDIAAACAFLVSEEAGYITGQILGVNGGRNT--
MAB_0833|M.abscessus_ATCC_1997 PDEMVGTALLLTSDAGSYITGQTIIADGGLVAR-
MAV_2438|M.avium_104 PEEVASITVYLASRRNGYVTGAMVNVDGGSDFI-
MMAR_1568|M.marinum_M PEEVASITAYLVSRRNGYVTGATVNADGGSDFI-
Mflv_4353|M.gilvum_PYR-GCK PTEIGSVVAFLASRRNSYMTGADINVDGGSDFT-
Mvan_1991|M.vanbaalenii_PYR-1 PTEIGPVVAFLASRRNSYMTGANINVDGGSDFT-
TH_4608|M.thermoresistible__bu PEEIGPVIAFLASRRNSYMTGANVNVDGGSDFV-
MSMEG_2228|M.smegmatis_MC2_155 PSEIGPVAAFLASRRNSYMTGANINVDGGSDFT-
:: *.* .*::* : . **