For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTASSAQMPPQSARQRVLDVALALFTRHGYAGTSLQMIADEMGVTKGSLYYHFRAREDLLRALVGPLIE QLRAIVEHAEAQRSPSVRAEHVLTGYARLAAANRALVAVTSGDPSVVEVIHADPDWADLIDRQIGLLAAV QPGLAGRVRATIVMAGIAATAAPGVVEADEAELCRQLIDAGRRTLGLRAPRA
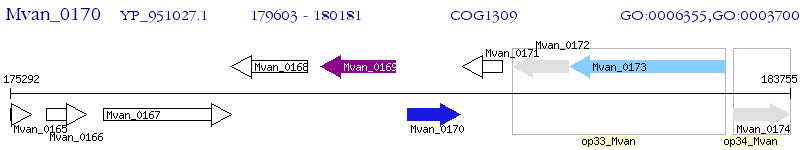
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0170 | - | - | 100% (192) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1613 | - | 5e-24 | 34.73% (167) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4249 | - | 5e-19 | 35.05% (194) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3185c | - | 3e-07 | 35.23% (88) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0973 | - | 7e-46 | 49.44% (178) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3160c | - | 3e-07 | 35.23% (88) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02568 | - | 3e-08 | 30.77% (104) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2677c | - | 4e-13 | 29.22% (154) | putative trancscriptional regulator,TetR |
| M. marinum M | MMAR_4699 | - | 6e-19 | 34.94% (166) | transcriptional regulatory protein |
| M. avium 104 | MAV_0959 | - | 8e-22 | 35.43% (175) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6537 | - | 9e-50 | 53.07% (179) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2336 | - | 2e-51 | 51.56% (192) | transcriptional regulator, TetR family protein |
| M. ulcerans Agy99 | MUL_0293 | - | 3e-16 | 48.28% (87) | transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3185c|M.bovis_AF2122/97 -----------------MPRQAGRWSPTALRILGAAAELIALRGYSSTST
Rv3160c|M.tuberculosis_H37Rv -----------------MPRQAGRWSPTALRILGAAAELIALRGYSSTST
Mflv_0973|M.gilvum_PYR-GCK --------------MSGDSTRAGGP--TRRRLIDVAIELFKRYSVAGTSL
TH_2336|M.thermoresistible__bu VSGVGKPISREAALLTSRASAGTGP--TRQRLIEVAIELFKQHSVAGTSL
MSMEG_6537|M.smegmatis_MC2_155 ---------------MDTSTANAAA--TRVRLVDAAVRLITRHGFAGTSL
Mvan_0170|M.vanbaalenii_PYR-1 --------------MTTASSAQMPPQSARQRVLDVALALFTRHGYAGTSL
MMAR_4699|M.marinum_M -------MSSQ----SRNQFRVITRSAAQVRILDATLGLIAEHGVGGTSL
MAV_0959|M.avium_104 -------MGTARQAVSDEQFWLVEHSPARTRVLDAALDLFAANGVSGTSL
MAB_2677c|M.abscessus_ATCC_199 -----------------MLSDAAVAPGTRERIQTVARDLFAERGLRNTSL
MUL_0293|M.ulcerans_Agy99 -----------------MVTKPRSD--TRRRIQEVARELFLRQGVRGTSL
MLBr_02568|M.leprae_Br4923 ---------------MAGGAKRIPRAVREQQMLDAAVQMFSVNGYHKTSM
:: .: :: . **
Mb3185c|M.bovis_AF2122/97 RDIAAAVGVEQPAIYKHFSAKRDILAALVRLAVEWPLELFGHITAMPVPA
Rv3160c|M.tuberculosis_H37Rv RDIAAAVGVEQPAIYKHFSAKRDILAALVRLAVEWPLELFGHITAMPVPA
Mflv_0973|M.gilvum_PYR-GCK QMISDELGLTKSAIYHHFRTRDELLTAVMEPLIAEVAEIVDAAEEQRTPH
TH_2336|M.thermoresistible__bu QMISDALGLTKSAIYHHFRTRDELLTAVMEPLISEVAEIVSAAEARRGAR
MSMEG_6537|M.smegmatis_MC2_155 QMIADELGFTKAAIYYHFRTREQLLGAVVEPFLSELQTVIDDAERLRGVH
Mvan_0170|M.vanbaalenii_PYR-1 QMIADEMGVTKGSLYYHFRAREDLLRALVGPLIEQLRAIVEHAEAQRSPS
MMAR_4699|M.marinum_M QMIADAVGVTKAAVYHQFKTKEQIVIALTERELGGLEEALEAAEAHDHPS
MAV_0959|M.avium_104 QMIADAVGITKAAVYHQFRTKEQIVIAVTERELGRLVPALEEAEAHDDGP
MAB_2677c|M.abscessus_ATCC_199 QDIASRLKITKPALYYHFSSRDELVRSIVQPMVDDLQTFLATNN---AGD
MUL_0293|M.ulcerans_Agy99 QDIADRLGITKPALYYHFASREDLVRSIVVPLIEEGERFVAERERLADGN
MLBr_02568|M.leprae_Br4923 DAIAAEAQISKPMLYLYYGSKEDLFGACLNREMSRFIAMVRADIDFTQSP
*: . : :* : :: ::. : : .
Mb3185c|M.bovis_AF2122/97 VVKLHRWLTESLDHLHASPYVLVSILITPDLHQESFVAERELVAEMERAL
Rv3160c|M.tuberculosis_H37Rv VVKLHRWLTESLDHLHASPYVLVSILITPDLHQESFVAERELVAEMERAL
Mflv_0973|M.gilvum_PYR-GCK ARAD--HMLVGYASIAARHRTLIPLLSGDPGVVALLRTRSEWGDLVRRQL
TH_2336|M.thermoresistible__bu TRAD--HFLRGYAALAAAHREVISLLTGDPGVVALLRTRPDWAEVVQRQM
MSMEG_6537|M.smegmatis_MC2_155 ARAD--HMVRGYAGLAVRNRALVSVLAGDPSVNEALKTRPEWAALIDRQM
Mvan_0170|M.vanbaalenii_PYR-1 VRAE--HVLTGYARLAAANRALVAVTSGDPSVVEVIHADPDWADLIDRQI
MMAR_4699|M.marinum_M QARE--LLLDRVIELAVERRGAASTLQFDPVIVRLLAEHQPFQRFIERLY
MAV_0959|M.avium_104 QARD--ALLVRVIEMAVRDRRLVRTLQFDPVVVRLLAEHEPFQRFMDRLY
MAB_2677c|M.abscessus_ATCC_199 RR----QLLEDYFDVLWTHRVVLGIIVRDLAALGQLELGEWMMGWRRRLI
MUL_0293|M.ulcerans_Agy99 VR----GLLEGYFDFHYRHRQDLMLVVVELTLLSDLGLIDTVLARRERLG
MLBr_02568|M.leprae_Br4923 KDLLRNTIVSFLRYIDANRASWIVMYIQAMSLPAFAQTVREARDQITELV
. . .
Mb3185c|M.bovis_AF2122/97 VGLIETGQGEGDVRAMHPLSAARLVQALFDALALPEFAVSPDEIVEFAMT
Rv3160c|M.tuberculosis_H37Rv VGLIETGQGEGDVRAMHPLSAARLVQALFDALALPEFAVSPDEIVEFAMT
Mflv_0973|M.gilvum_PYR-GCK ALFSEVEPGLGGQVKAALAMSGIASAAGVDYGGDVDDDTLRDQLIAAGRR
TH_2336|M.thermoresistible__bu ALLGDVQPGPGGRVKAAVVMAGIAAAAGVDYG-DLDEAALRDELLAAGRR
MSMEG_6537|M.smegmatis_MC2_155 RLLTDVAPGPAGRVKAMMVFSGMSAAAGPASD-DIPVEELYAHLVDTSRR
Mvan_0170|M.vanbaalenii_PYR-1 GLLAAVQPGLAGRVRATIVMAGIAATAAPGVV-EADEAELCRQLIDAGRR
MMAR_4699|M.marinum_M GVLIG-DAGQDTRVLAAMLSGAIAVGVLHPLVADVDDATLRTQLRRITSR
MAV_0959|M.avium_104 RVLLS-DAGLDGRIEAAMFSGALSTAVMHPLVVDIDDETLLDRVTDLSRR
MAB_2677c|M.abscessus_ATCC_199 VLLAGGKPSAAQMIQATVALGGLSDCTIEYAD--RPHRQVKKTAVAAAIA
MUL_0293|M.ulcerans_Agy99 RLVFGAAPTLGQATRAVLAFGGLQDCCMQFPD--EPYAQLRQYSVEAALA
MLBr_02568|M.leprae_Br4923 AGLMRVGTRSSRPDHEYEIMAGALVGAGEAMATRLTTGDIAVDEAAELMI
. ..
Mb3185c|M.bovis_AF2122/97 ALLSDPDRLAEIRAAADALEIQTAPPDRGL
Rv3160c|M.tuberculosis_H37Rv ALLSDPDRLAEIRAAADALEIQTAPPDRGL
Mflv_0973|M.gilvum_PYR-GCK ALGLRNSRTGAVTARR--------------
TH_2336|M.thermoresistible__bu TLGLR-----APHARG--------------
MSMEG_6537|M.smegmatis_MC2_155 ALGLR-----APREKA--------------
Mvan_0170|M.vanbaalenii_PYR-1 TLGLR-----APRA----------------
MMAR_4699|M.marinum_M LMDTGE----G-------------------
MAV_0959|M.avium_104 LLGLPR----KPVE----------------
MAB_2677c|M.abscessus_ATCC_199 ALES--------------------------
MUL_0293|M.ulcerans_Agy99 ALGV--------------------------
MLBr_02568|M.leprae_Br4923 NLFWHGLKGAPADRDFGPSIVAG-------
: