For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSDAAVAPGTRERIQTVARDLFAERGLRNTSLQDIASRLKITKPALYYHFSSRDELVRSIVQPMVDDLQT FLATNNAGDRRQLLEDYFDVLWTHRVVLGIIVRDLAALGQLELGEWMMGWRRRLIVLLAGGKPSAAQMIQ ATVALGGLSDCTIEYADRPHRQVKKTAVAAAIAALES*
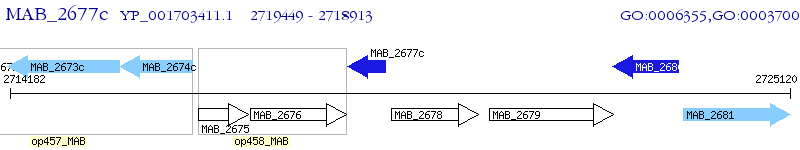
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2677c | - | - | 100% (178) | putative trancscriptional regulator,TetR |
| M. abscessus ATCC 19977 | MAB_4912c | - | 3e-11 | 35.19% (108) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0599 | - | 3e-08 | 36.07% (61) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1047 | - | 9e-08 | 36.00% (75) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0973 | - | 4e-12 | 25.71% (175) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1019 | - | 9e-08 | 36.00% (75) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00815 | - | 3e-07 | 28.74% (87) | putative TetR-family transcriptional regulator |
| M. marinum M | MMAR_4674 | - | 2e-39 | 48.52% (169) | transcriptional regulator |
| M. avium 104 | MAV_0985 | - | 4e-33 | 44.74% (152) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_5718 | - | 2e-40 | 50.00% (172) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2336 | - | 2e-14 | 26.97% (178) | transcriptional regulator, TetR family protein |
| M. ulcerans Agy99 | MUL_0293 | - | 5e-38 | 47.93% (169) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1613 | - | 2e-14 | 29.38% (160) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4674|M.marinum_M --------------------------------MTKPRSDTRRRIQEVARE
MUL_0293|M.ulcerans_Agy99 -------------------------------MVTKPRSDTRRRIQEVARE
MAV_0985|M.avium_104 --------------------------------------------------
MSMEG_5718|M.smegmatis_MC2_155 -------------------------------------MDTRQRIQEVARE
MAB_2677c|M.abscessus_ATCC_199 -----------------------------MLSDAAVAPGTRERIQTVARD
Mflv_0973|M.gilvum_PYR-GCK ----------------------------MSGDSTRAGGPTRRRLIDVAIE
TH_2336|M.thermoresistible__bu --------------VSGVGKPISREAALLTSRASAGTGPTRQRLIEVAIE
Mb1047|M.bovis_AF2122/97 ----------------------------------MTGTERRHQLIGIARS
Rv1019|M.tuberculosis_H37Rv ----------------------------------MTGTERRHQLIGIARS
MLBr_00815|M.leprae_Br4923 MNNLAKAAQRRTVRPIDRGRLSDDIATPNQRSNRLSRDKRRGQLLIVASD
Mvan_1613|M.vanbaalenii_PYR-1 -----------------------------MAKPVATRGFARERVLESALE
MMAR_4674|M.marinum_M LFLRQGVRGTSLQDIADRLGITKPALYYHFASREDLVRSIVVPLIEEGER
MUL_0293|M.ulcerans_Agy99 LFLRQGVRGTSLQDIADRLGITKPALYYHFASREDLVRSIVVPLIEEGER
MAV_0985|M.avium_104 ------MQRTSLQDIADRLGITKPALYYHFPSREDLVRSILVPLIEEGER
MSMEG_5718|M.smegmatis_MC2_155 LFAEKGVVRTSLQDIADRLGITKPALYYHFRSREELIRSILQPLIDEGEA
MAB_2677c|M.abscessus_ATCC_199 LFAERGLRNTSLQDIASRLKITKPALYYHFSSRDELVRSIVQPMVDDLQT
Mflv_0973|M.gilvum_PYR-GCK LFKRYSVAGTSLQMISDELGLTKSAIYHHFRTRDELLTAVMEPLIAEVAE
TH_2336|M.thermoresistible__bu LFKQHSVAGTSLQMISDALGLTKSAIYHHFRTRDELLTAVMEPLISEVAE
Mb1047|M.bovis_AF2122/97 LFAERGYDGTSIEEIAQRANVSKPVVYEHFGGKEGLYAVVVDREMSALLD
Rv1019|M.tuberculosis_H37Rv LFAERGYDGTSIEEIAQRANVSKPVVYEHFGGKEGLYAVVVDREMSALLD
MLBr_00815|M.leprae_Br4923 VFVDHGYHAAGMDEIADRAGVSKPVLYQHFSSKLELYLAVLERHMNNLVS
Mvan_1613|M.vanbaalenii_PYR-1 LFADHGVSGTSLQMIADRLGVTKAAVYYQFHSKDEITLAVVKPVFDDIEH
:.:: *:. ::*..:* :* : : :: .
MMAR_4674|M.marinum_M FVAERERL---ADGNVRGLLEGYFDFHYRHRQDLMLVVVE-LTLLSDLGL
MUL_0293|M.ulcerans_Agy99 FVAERERL---ADGNVRGLLEGYFDFHYRHRQDLMLVVVE-LTLLSDLGL
MAV_0985|M.avium_104 FVAEHESR---EHTEARELLEGYFDFHYRHRRDLVLLLTE-LSTLIDLDL
MSMEG_5718|M.smegmatis_MC2_155 FVAEQERKRAADRASARELLEGYFDFHYRHRADLVLIVSE-LTTLVDLGL
MAB_2677c|M.abscessus_ATCC_199 FLATNNAG------DRRQLLEDYFDVLWTHRVVLGIIVRD-LAALGQLEL
Mflv_0973|M.gilvum_PYR-GCK IVDAAEEQR-TPHARADHMLVGYASIAARHRTLIPLLSGD-PGVVALLRT
TH_2336|M.thermoresistible__bu IVSAAEARR-GARTRADHFLRGYAALAAAHREVISLLTGD-PGVVALLRT
Mb1047|M.bovis_AF2122/97 GITSSLTNN-RSRVRVERVALALLTYVEERTDGFRIMIRDSPASISSGTY
Rv1019|M.tuberculosis_H37Rv GITSSLTNN-RSRVRVERVALALLTYVEERTDGFRIMIRDSPASISSGTY
MLBr_00815|M.leprae_Br4923 SVQQALRTTTNNRQRLHAAVQAFFDFIEHDGQGYRLIFEN-DNITEPRVA
Mvan_1613|M.vanbaalenii_PYR-1 LLRITETLP-TPEARRSVSMSGLVELIIRHRRVSSVFYGD-PTVHHLLEG
: :. :
MMAR_4674|M.marinum_M IDTVLAWRERLGRLVFGAAPTLGQA-TRAVLAFGGLQDCC-MQFP-DEPY
MUL_0293|M.ulcerans_Agy99 IDTVLARRERLGRLVFGAAPTLGQA-TRAVLAFGGLQDCC-MQFP-DEPY
MAV_0985|M.avium_104 IDTVLAWRERLARLVFGKRPTLAQS-TRAVVAFGGLQDCC-VQFP-DAPQ
MSMEG_5718|M.smegmatis_MC2_155 IDQLLAWRARLGALVFGRTPTLEQS-TRAVIAFGGLQDCC-LQFP-DVPA
MAB_2677c|M.abscessus_ATCC_199 GEWMMGWRRRLIVLLAGGKPSAAQM-IQATVALGGLSDCT-IEYA-DRPH
Mflv_0973|M.gilvum_PYR-GCK RSEWGDLVRRQLALFSEVEPGLGGQ-VKAALAMSGIASAAGVDYGGDVDD
TH_2336|M.thermoresistible__bu RPDWAEVVQRQMALLGDVQPGPGGR-VKAAVVMAGIAAAAGVDYG-DLDE
Mb1047|M.bovis_AF2122/97 SSLLNDAVSQVSSILAGDFARRGLDPDLAPLYAQALVGSVSMTAQWWLDA
Rv1019|M.tuberculosis_H37Rv SSLLNDAVSQVSSILAGDFARRGLDPDLAPLYAQALVGSVSMTAQWWLDA
MLBr_00815|M.leprae_Br4923 AQVRVATESCIDAVFALISADSRLDPHRARMVAVGLVGISVDCARYWLDT
Mvan_1613|M.vanbaalenii_PYR-1 NDEFRRLGDRLADVLLGPDPDTATR-VAISMLSAGIHGCVTDPMIGDVAD
:. . : .:
MMAR_4674|M.marinum_M AQ-LRQYSVEAALAALGV---------------
MUL_0293|M.ulcerans_Agy99 AQ-LRQYSVEAALAALGV---------------
MAV_0985|M.avium_104 AE-LRDKSVAAALDALGERPRR-----------
MSMEG_5718|M.smegmatis_MC2_155 EQ-LRTATVNGAMAALGL---------------
MAB_2677c|M.abscessus_ATCC_199 RQ-VKKTAVAAAIAALES---------------
Mflv_0973|M.gilvum_PYR-GCK DT-LRDQLIAAGRRALGLRNSRTGAVTARR---
TH_2336|M.thermoresistible__bu AA-LRDELLAAGRRTLGLR-----APHARG---
Mb1047|M.bovis_AF2122/97 RE-PKKEVVAAHLVNLVWNGLTHLEADPRLQDE
Rv1019|M.tuberculosis_H37Rv RE-PKKEVVAAHLVNLVWNGLTHLEADPRLQDE
MLBr_00815|M.leprae_Br4923 NRPISKSDAVEGTVQFAWGGLSHVPLTRL----
Mvan_1613|M.vanbaalenii_PYR-1 AE-LQQILLDLTKAFLDVCVHD-----------
: