For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GADATKVLRPTVVCQDGHVRTRSKRWGGRTGAERRAERRQRLIDAATEIWSESGWAAVTMRGVCAKTGLN DRYFYEDFKTRDELLVAAWDGVRNEMLGEVSATLAERIGQPPMETIRVTIAAVVDRIAEDPGRAKILLAQ HVGSSPLQDRRAVALQEATHLVIAASEPHLRPEADETALRMDTLVAVGGFVELITAWHAGLLDVIQGEIV EHTSRLAETLAERYLVDDAGL*
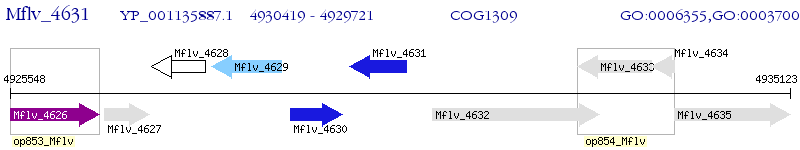
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4631 | - | - | 100% (232) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4611 | - | 8e-97 | 77.23% (224) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1526 | - | 9e-22 | 32.40% (179) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0281c | - | 6e-15 | 28.17% (213) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0275c | - | 3e-14 | 27.70% (213) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2070c | - | 1e-95 | 80.48% (210) | putative transcriptional regulator |
| M. marinum M | MMAR_3142 | - | 4e-96 | 81.73% (208) | TetR family transcriptional regulator |
| M. avium 104 | MAV_3986 | - | 6e-25 | 35.42% (192) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1355 | - | 1e-24 | 34.18% (196) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_4000 | - | 2e-23 | 34.48% (203) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_1200 | - | 8e-18 | 28.86% (201) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1244 | - | 3e-22 | 32.81% (192) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1355|M.smegmatis_MC2_155 --------------------MTTPTRWAGVPLTDRRAERRALLVDAAFRL
TH_4000|M.thermoresistible__bu --------------------MTSPTRWAGVPLTDRRAERRALLIAAAYRL
Mvan_1244|M.vanbaalenii_PYR-1 ------------MSSAASRPAATPTRWAGVPLTDRRAERRALLVDAAFRL
MAV_3986|M.avium_104 --------------------MSSPSRWAGVPLKDRRAERRALLVEAAYRL
Mb0281c|M.bovis_AF2122/97 -------------------MTRSDRPYRGVEAAERLATRRRQLLSAGLDL
Rv0275c|M.tuberculosis_H37Rv -------------------MTRSDRPYRGVEAAERLATRRRQSLSAGLDL
MUL_1200|M.ulcerans_Agy99 -----------------------MRPYRGVEATERLATRRSQLLESGLEL
MAB_2070c|M.abscessus_ATCC_199 -------------------MRSRNKRWGGRTGAERRAERRQQLIEAATEI
MMAR_3142|M.marinum_M ----------------MFDVRSRNTRWGGRTGAERRAERRQQLIEAATEI
Mflv_4631|M.gilvum_PYR-GCK MGADATKVLRPTVVCQDGHVRTRSKRWGGRTGAERRAERRQRLIDAATEI
: * :* * ** : :. :
MSMEG_1355|M.smegmatis_MC2_155 FGDGGE--TALSVRSVCRECGLNTRYFYESFADTDDLIGAVYDEVSALLA
TH_4000|M.thermoresistible__bu FGEGGE--AALSVRSVCRESELNSRYFYESFADVDELLGAVYDEVSAQLA
Mvan_1244|M.vanbaalenii_PYR-1 FGDGGE--QALSVRSVSREAGLNTRYFYESFTDTDDLLGAVYDRVSAELA
MAV_3986|M.avium_104 FGSEGE--AAVSVRSVCRECGLNTRYFYESFRDTDDLLGAVYDRVSEQLE
Mb0281c|M.bovis_AF2122/97 LGSDQHDIAELTIRTICRRAGLSVRYFYESFTDKDEFVGRVFDWVVAELV
Rv0275c|M.tuberculosis_H37Rv LGSDQHDIAELTIRTICRRAGLSVRYFYESFTDKDEFVGRVFDWVVAELV
MUL_1200|M.ulcerans_Agy99 LGSSDRDIAELTVRGICRQAGLSARYFYESFPDKDEFIGGVFDWVVGDLA
MAB_2070c|M.abscessus_ATCC_199 WSESGW--AAVTMRGVCARTSLNDRYFYEDFKTRDELLVAAWDGVRNDML
MMAR_3142|M.marinum_M WSESGW--AAVTMRGVCARTGLNDRYFYEDFKTREDLLVAAWDGVRNDML
Mflv_4631|M.gilvum_PYR-GCK WSESGW--AAVTMRGVCAKTGLNDRYFYEDFKTRDELLVAAWDGVRNEML
.. :::* :. . *. *****.* :::: .:* * :
MSMEG_1355|M.smegmatis_MC2_155 ADVEAAM--------EGAGNSLRARTRAGIAAVLGFSSADPRRGRVLFTD
TH_4000|M.thermoresistible__bu AEVETAMAAAVEGAGEGAGDSLRARTRAGIVAVLGFSSADPRRGRILFTD
Mvan_1244|M.vanbaalenii_PYR-1 EVVEAAMS--------GAGASLPARTRAGMTAVLHFSSADPRRGRVLFTD
MAV_3986|M.avium_104 EVIAAAIE--------QAGDSLRARTRAGIAAVLGFSSADPRRGRVLFTD
Mb0281c|M.bovis_AF2122/97 ATTQAAVT----------AVPAREQTRAGMANIVRTITADARVGRLLFST
Rv0275c|M.tuberculosis_H37Rv ATTQAAVT----------AVPAREQTRAGMANIVRTITADARVGRLLFST
MUL_1200|M.ulcerans_Agy99 TTIQAAVA----------TVPPPEQARAGIAEIVRTIVGDARIGRLLFST
MAB_2070c|M.abscessus_ATCC_199 GEVAALFTER-------ANRPPIETITAAIAIVVDGIARDPGRAHILLAQ
MMAR_3142|M.marinum_M GEVSALFDER-------VDRPPIETITAAIAIVVDRIARDPGRAHILLAQ
Mflv_4631|M.gilvum_PYR-GCK GEVSATLAER-------IGQPPMETIRVTIAAVVDRIAEDPGRAKILLAQ
: . . . :. :: *. .::*::
MSMEG_1355|M.smegmatis_MC2_155 ARANPVLAQRRAVTQDLLREAVLSEGG-RLNPGSDPVAAQVGAAMYTGAM
TH_4000|M.thermoresistible__bu ARANPVLAARRAATQDQLREAVLTEGW-RLHPKSDPVAAQVGAAMYTGAM
Mvan_1244|M.vanbaalenii_PYR-1 ARANPVLTQRRAATQDLLREAVLTEGW-RLHPDSDPTAAMVGAAMYTGAM
MAV_3986|M.avium_104 ARTNPVLAARRRATQDMLRQGVLSEGW-RLNPRSDPVAAEVAAAMYTGAM
Mb0281c|M.bovis_AF2122/97 QLANAVITRKRAESSALFAMLSGQHAV-DTLHAPANDHVKAVAHFAVGGV
Rv0275c|M.tuberculosis_H37Rv QLANAVITRKRAESSALFAMLSGQHAV-DTLHAPANDHVKAVAHFAVGGV
MUL_1200|M.ulcerans_Agy99 HLANPVVVRKRTESSALFAMLSGQHAG-DALRIPANDRIKVGAHFIVGGV
MAB_2070c|M.abscessus_ATCC_199 HVGSSPLQDRRAVALQEATQLVVEASRPHLRHDADEMALRMDTLIAVGGF
MMAR_3142|M.marinum_M HVGSSPLQDRRAVALQEATQLVVEASRPHLREDADETALRMDTLVAVGGF
Mflv_4631|M.gilvum_PYR-GCK HVGSSPLQDRRAVALQEATHLVIAASEPHLRPEADETALRMDTLVAVGGF
.. : :* : . . : . .*..
MSMEG_1355|M.smegmatis_MC2_155 AELAQQWLAGNLGDDLDAVVDHALRLVLGR--------------------
TH_4000|M.thermoresistible__bu AELAQQWLAGNLGDDLDRVVDYALRLVLGR--------------------
Mvan_1244|M.vanbaalenii_PYR-1 AELAQQWLAGHLGDDLDAVVGYALRLVLR---------------------
MAV_3986|M.avium_104 AELAQQWLAGHLGGDLDAVVDYASKLVLR---------------------
Mb0281c|M.bovis_AF2122/97 GQTISAWLAGDVRLDPDQLVDQLAALLDELTDPNLSRPRVAATAAKSGAN
Rv0275c|M.tuberculosis_H37Rv GQTISAWLAGDVRLDPDQLVDQLAALLDELTDPNLSRPRVAATAAKSGAN
MUL_1200|M.ulcerans_Agy99 AQTISAWIEGDVELEPGQLVDQLASLLDELSDPRLYGLR----GTRAGAT
MAB_2070c|M.abscessus_ATCC_199 VEVITAWHSGLLAVTEKEVVAHTSRLAETLAQRYVVSD------------
MMAR_3142|M.marinum_M VEVITAWHSGLLAVTEKEVVAHTSRLAETLAQRYVVSG------------
Mflv_4631|M.gilvum_PYR-GCK VELITAWHAGLLDVIQGEIVEHTSRLAETLAERYLVDDAGL---------
: * * : :* *
MSMEG_1355|M.smegmatis_MC2_155 ---------------------
TH_4000|M.thermoresistible__bu ---------------------
Mvan_1244|M.vanbaalenii_PYR-1 ---------------------
MAV_3986|M.avium_104 ---------------------
Mb0281c|M.bovis_AF2122/97 DPQPPEVAGQPPSSARPARRS
Rv0275c|M.tuberculosis_H37Rv DPQPPEVAGQPPSSARPARRS
MUL_1200|M.ulcerans_Agy99 AASPAQ---------------
MAB_2070c|M.abscessus_ATCC_199 ---------------------
MMAR_3142|M.marinum_M ---------------------
Mflv_4631|M.gilvum_PYR-GCK ---------------------