For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRRSCLCQDVFVRTKAARWGGRTGAERRADRRRRLVNAATEIWTESGWAAVTMRGVCSRAALNDRYFYEE FKTRDELLVAAWDNVRDEMLGEVSATFAERVGQPPIETIRAAISIVVLRIAQDAGRAQILLTQHVGSSTL QDRRAVALQEATQLVMAASKPHLKPDADEMALRMDTFVAVGGFVELISAWHAGLLVDVSQVDIVEHTSRL AETLARRYVVDKFG
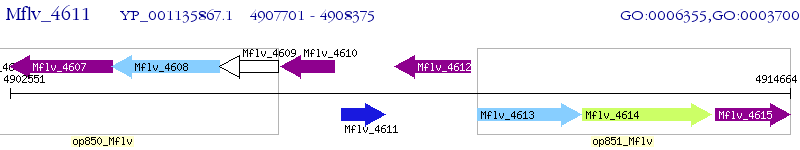
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4611 | - | - | 100% (224) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4631 | - | 8e-97 | 77.23% (224) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1526 | - | 1e-19 | 30.51% (177) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0281c | - | 8e-15 | 31.69% (183) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0275c | - | 4e-14 | 31.15% (183) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2070c | - | 3e-88 | 75.60% (209) | putative transcriptional regulator |
| M. marinum M | MMAR_3142 | - | 4e-88 | 76.08% (209) | TetR family transcriptional regulator |
| M. avium 104 | MAV_3986 | - | 5e-21 | 35.75% (179) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1355 | - | 8e-22 | 32.99% (197) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_4000 | - | 1e-21 | 32.98% (191) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_1200 | - | 5e-15 | 29.35% (201) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1244 | - | 2e-20 | 34.44% (180) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1355|M.smegmatis_MC2_155 ------------MTTPTRWAGVPLTDRRAERRALLVDAAFRLFGDGGE--
TH_4000|M.thermoresistible__bu ------------MTSPTRWAGVPLTDRRAERRALLIAAAYRLFGEGGE--
Mvan_1244|M.vanbaalenii_PYR-1 ----MSSAASRPAATPTRWAGVPLTDRRAERRALLVDAAFRLFGDGGE--
MAV_3986|M.avium_104 ------------MSSPSRWAGVPLKDRRAERRALLVEAAYRLFGSEGE--
Mb0281c|M.bovis_AF2122/97 -----------MTRSDRPYRGVEAAERLATRRRQLLSAGLDLLGSDQHDI
Rv0275c|M.tuberculosis_H37Rv -----------MTRSDRPYRGVEAAERLATRRRQSLSAGLDLLGSDQHDI
MUL_1200|M.ulcerans_Agy99 ---------------MRPYRGVEATERLATRRSQLLESGLELLGSSDRDI
MAB_2070c|M.abscessus_ATCC_199 -----------MRSRNKRWGGRTGAERRAERRQQLIEAATEIWSESGW--
MMAR_3142|M.marinum_M --------MFDVRSRNTRWGGRTGAERRAERRQQLIEAATEIWSESGW--
Mflv_4611|M.gilvum_PYR-GCK MRRSCLCQDVFVRTKAARWGGRTGAERRADRRRRLVNAATEIWTESGW--
: * :* * ** : :. : .
MSMEG_1355|M.smegmatis_MC2_155 TALSVRSVCRECGLNTRYFYESFADTDDLIGAVYDEVSALLAADVEAAM-
TH_4000|M.thermoresistible__bu AALSVRSVCRESELNSRYFYESFADVDELLGAVYDEVSAQLAAEVETAMA
Mvan_1244|M.vanbaalenii_PYR-1 QALSVRSVSREAGLNTRYFYESFTDTDDLLGAVYDRVSAELAEVVEAAMS
MAV_3986|M.avium_104 AAVSVRSVCRECGLNTRYFYESFRDTDDLLGAVYDRVSEQLEEVIAAAIE
Mb0281c|M.bovis_AF2122/97 AELTIRTICRRAGLSVRYFYESFTDKDEFVGRVFDWVVAELVATTQAAVT
Rv0275c|M.tuberculosis_H37Rv AELTIRTICRRAGLSVRYFYESFTDKDEFVGRVFDWVVAELVATTQAAVT
MUL_1200|M.ulcerans_Agy99 AELTVRGICRQAGLSARYFYESFPDKDEFIGGVFDWVVGDLATTIQAAVA
MAB_2070c|M.abscessus_ATCC_199 AAVTMRGVCARTSLNDRYFYEDFKTRDELLVAAWDGVRNDMLGEVAALFT
MMAR_3142|M.marinum_M AAVTMRGVCARTGLNDRYFYEDFKTREDLLVAAWDGVRNDMLGEVSALFD
Mflv_4611|M.gilvum_PYR-GCK AAVTMRGVCSRAALNDRYFYEEFKTRDELLVAAWDNVRDEMLGEVSATFA
:::* :. . *. *****.* :::: .:* * : : .
MSMEG_1355|M.smegmatis_MC2_155 -------EGAGNSLRARTRAGIAAVLGFSSADPRRGRVLFTDARANPVLA
TH_4000|M.thermoresistible__bu AAVEGAGEGAGDSLRARTRAGIVAVLGFSSADPRRGRILFTDARANPVLA
Mvan_1244|M.vanbaalenii_PYR-1 --------GAGASLPARTRAGMTAVLHFSSADPRRGRVLFTDARANPVLT
MAV_3986|M.avium_104 --------QAGDSLRARTRAGIAAVLGFSSADPRRGRVLFTDARTNPVLA
Mb0281c|M.bovis_AF2122/97 ----------AVPAREQTRAGMANIVRTITADARVGRLLFSTQLANAVIT
Rv0275c|M.tuberculosis_H37Rv ----------AVPAREQTRAGMANIVRTITADARVGRLLFSTQLANAVIT
MUL_1200|M.ulcerans_Agy99 ----------TVPPPEQARAGIAEIVRTIVGDARIGRLLFSTHLANPVVV
MAB_2070c|M.abscessus_ATCC_199 ER-------ANRPPIETITAAIAIVVDGIARDPGRAHILLAQHVGSSPLQ
MMAR_3142|M.marinum_M ER-------VDRPPIETITAAIAIVVDRIARDPGRAHILLAQHVGSSPLQ
Mflv_4611|M.gilvum_PYR-GCK ER-------VGQPPIETIRAAISIVVLRIAQDAGRAQILLTQHVGSSTLQ
. *.: :: *. .::*:: .. :
MSMEG_1355|M.smegmatis_MC2_155 QRRAVTQDLLREAVLSEGG-RLNPGSDPVAAQVGAAMYTGAMAELAQQWL
TH_4000|M.thermoresistible__bu ARRAATQDQLREAVLTEGW-RLHPKSDPVAAQVGAAMYTGAMAELAQQWL
Mvan_1244|M.vanbaalenii_PYR-1 QRRAATQDLLREAVLTEGW-RLHPDSDPTAAMVGAAMYTGAMAELAQQWL
MAV_3986|M.avium_104 ARRRATQDMLRQGVLSEGW-RLNPRSDPVAAEVAAAMYTGAMAELAQQWL
Mb0281c|M.bovis_AF2122/97 RKRAESSALFAMLSGQHAV-DTLHAPANDHVKAVAHFAVGGVGQTISAWL
Rv0275c|M.tuberculosis_H37Rv RKRAESSALFAMLSGQHAV-DTLHAPANDHVKAVAHFAVGGVGQTISAWL
MUL_1200|M.ulcerans_Agy99 RKRTESSALFAMLSGQHAG-DALRIPANDRIKVGAHFIVGGVAQTISAWI
MAB_2070c|M.abscessus_ATCC_199 DRRAVALQEATQLVVEASRPHLRHDADEMALRMDTLIAVGGFVEVITAWH
MMAR_3142|M.marinum_M DRRAVALQEATQLVVEASRPHLREDADETALRMDTLVAVGGFVEVITAWH
Mflv_4611|M.gilvum_PYR-GCK DRRAVALQEATQLVMAASKPHLKPDADEMALRMDTFVAVGGFVELISAWH
:* : . . : . .*.. : *
MSMEG_1355|M.smegmatis_MC2_155 AGNLG-DDLDAVVDHALRLVLGR---------------------------
TH_4000|M.thermoresistible__bu AGNLG-DDLDRVVDYALRLVLGR---------------------------
Mvan_1244|M.vanbaalenii_PYR-1 AGHLG-DDLDAVVGYALRLVLR----------------------------
MAV_3986|M.avium_104 AGHLG-GDLDAVVDYASKLVLR----------------------------
Mb0281c|M.bovis_AF2122/97 AGDVR-LDPDQLVDQLAALLDELTDPNLSRPRVAATAAKSGANDPQPPEV
Rv0275c|M.tuberculosis_H37Rv AGDVR-LDPDQLVDQLAALLDELTDPNLSRPRVAATAAKSGANDPQPPEV
MUL_1200|M.ulcerans_Agy99 EGDVE-LEPGQLVDQLASLLDELSDPRLYGLR----GTRAGATAASPAQ-
MAB_2070c|M.abscessus_ATCC_199 SGLLA-VTEKEVVAHTSRLAETLAQRYVVSD-------------------
MMAR_3142|M.marinum_M SGLLA-VTEKEVVAHTSRLAETLAQRYVVSG-------------------
Mflv_4611|M.gilvum_PYR-GCK AGLLVDVSQVDIVEHTSRLAETLARRYVVDKFG-----------------
* : :* *
MSMEG_1355|M.smegmatis_MC2_155 --------------
TH_4000|M.thermoresistible__bu --------------
Mvan_1244|M.vanbaalenii_PYR-1 --------------
MAV_3986|M.avium_104 --------------
Mb0281c|M.bovis_AF2122/97 AGQPPSSARPARRS
Rv0275c|M.tuberculosis_H37Rv AGQPPSSARPARRS
MUL_1200|M.ulcerans_Agy99 --------------
MAB_2070c|M.abscessus_ATCC_199 --------------
MMAR_3142|M.marinum_M --------------
Mflv_4611|M.gilvum_PYR-GCK --------------