For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SARISVIGAGISGLAAAVALERAGHHVSVIEQRTDTGAGSGISLWPNALAALDQIGLGDSVRDAGGRVTA GAIRWRDGTWVRRPSVRRIVDALGEPLVVVRRSRLTDILREALPAGTVRTGLAATRVSVGASSVRITLSD GEVRESDAVVGADGVNSVLARTLNGPLRTRYVGYTAWRGIAAHPLDPELGGETLGPGTQVGHVPLGPDHT YWFATERTAEGGSAPGGEHAYLTAKVADWADPIPRLVATTDPGDLLRNDLYDRARAARWSDGRAVLIGDA AHPMRPHLGQGGCQGIEDAAILARFLELADDVPTAFERFEAFRKPRVGMLVREAQTLGRIVNVRPAVLSG LAGRATALIPERMVTAHLAWIAAGSAFVLPTAADLAR*
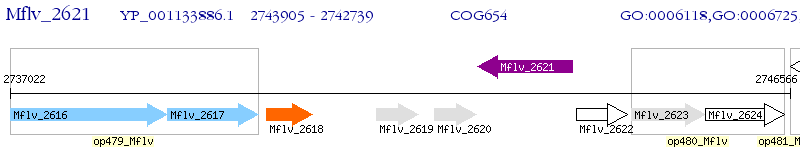
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2621 | - | - | 100% (388) | monooxygenase, FAD-binding |
| M. gilvum PYR-GCK | Mflv_0304 | - | 3e-08 | 25.25% (396) | hypothetical protein Mflv_0304 |
| M. gilvum PYR-GCK | Mflv_5254 | - | 1e-07 | 26.97% (330) | geranylgeranyl reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0590c | - | 2e-12 | 26.98% (341) | hypothetical protein Mb0590c |
| M. tuberculosis H37Rv | Rv0575c | - | 2e-12 | 26.98% (341) | hypothetical protein Rv0575c |
| M. leprae Br4923 | MLBr_02276 | - | 2e-09 | 26.21% (351) | putative FAD-linked oxidoreductase |
| M. abscessus ATCC 19977 | MAB_1601c | - | 2e-98 | 47.66% (384) | putative monooxygenase |
| M. marinum M | MMAR_4944 | - | 3e-38 | 29.80% (349) | oxidoreductase |
| M. avium 104 | MAV_4556 | - | 3e-32 | 68.60% (86) | monooxygenase |
| M. smegmatis MC2 155 | MSMEG_4641 | - | 1e-140 | 64.81% (378) | salicylate hydroxylase |
| M. thermoresistible (build 8) | TH_1880 | - | 9e-08 | 25.81% (310) | flavin-type hydroxylase |
| M. ulcerans Agy99 | MUL_2358 | - | 3e-22 | 28.31% (378) | FAD-dependent oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_3960 | - | 1e-165 | 73.06% (386) | monooxygenase, FAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0590c|M.bovis_AF2122/97 -------------MKVAISGAGVAGAALAHWLQRTGHTPTVIERAPKFRT
Rv0575c|M.tuberculosis_H37Rv -------------MKVAISGAGVAGAALAHWLQRTGHTPTVIERAPKFRT
MUL_2358|M.ulcerans_Agy99 -------------MRVAVCGAGIAGLALAERMSALGAEVVFVERSPGPPN
Mflv_2621|M.gilvum_PYR-GCK -----------MSARISVIGAGISGLAAAVALERAGHHVSVIEQRTD-TG
Mvan_3960|M.vanbaalenii_PYR-1 -----------MPAHTIVIGAGIAGLATAVALRRVGHDVTVIEQRTD-LT
MSMEG_4641|M.smegmatis_MC2_155 -----------MAASFLVVGAGITGLATAAALQRRGHDVCVAEARAD-TA
MAV_4556|M.avium_104 MPAPCGVRFDTMAQRILVIGAGIAGLATAVAMQRRGYAVTVVKERTD-TS
MAB_1601c|M.abscessus_ATCC_199 -----------MSDHIVVVGAGIAGLATAVAIQQSGRDVVVVDDRDG-TS
MMAR_4944|M.marinum_M -------------MKAVIIGAGIGGMSAAIALRQIGIDTEVYERVTENKP
TH_1880|M.thermoresistible__bu ------------VYDVIVVGGGPTGMMLAGELRLHGVRVLVVEKNSEPPA
MLBr_02276|M.leprae_Br4923 -------MRVETSADVVVVGAGPAGSAAATWAARAGNDVIVIDSVCFPRD
: *.* * * * . .
Mb0590c|M.bovis_AF2122/97 GGYMIDFWGVGYQVAKRMGITDQIAAAGYHMEHVRSVGPT-GKVKADLGV
Rv0575c|M.tuberculosis_H37Rv GGYMIDFWGVGYQVAKRMGITDQIAAAGYHMEHVRSVGPT-GKVKADLGV
MUL_2358|M.ulcerans_Agy99 QGAMIDLFGAGYEAADAIGILDAIRDVSYPIGDTGLVDEL-GRRRAGLPY
Mflv_2621|M.gilvum_PYR-GCK AGSGISLWPNALAALDQIGLGDSVRDAGGRVTAGAIRWRD-GTWVRRPSV
Mvan_3960|M.vanbaalenii_PYR-1 SGAGISIWPNALAALDQIGLGDNVRQAGGRVTAGAIRWRD-GTWLRRPSA
MSMEG_4641|M.smegmatis_MC2_155 SGAGISIWPNALAALDAIGLGDPVRAAGGRVTAGALRWHD-GTWLRHPAA
MAV_4556|M.avium_104 SGAGISIWPNALAALDRIGVGQAVRDAGGRITAGALRWRD-GSWLRRPAF
MAB_1601c|M.abscessus_ATCC_199 AGYAITLWPNALAACDALGIGEDVRAASARVEAGTMRWYD-GRILREPPS
MMAR_4944|M.marinum_M VGAAISVWSNGVKCLNYLGLEQQTARLGGIVETMSYAEAHSGETMCRISM
TH_1880|M.thermoresistible__bu HVRSLGLHVRSIEMLDQRGLLDRFLAHGQRHPVGGFFAAI------AKPP
MLBr_02276|M.leprae_Br4923 KACGDGLTPRAVAELKRLGLDDWLNTR---IRHRGLRMSGFGGEVEVDWP
. . . *: :
Mb0590c|M.bovis_AF2122/97 DVFRRMVGDDFTSLPRGDLAAAIYTTIEDQVETIFDDSIATIDEHRDGVR
Rv0575c|M.tuberculosis_H37Rv DVFRRMVGDDFTSLPRGDLAAAIYTTIEDQVETIFDDSIATIDEHRDGVR
MUL_2358|M.ulcerans_Agy99 GRIAKALEDRLCRITRTDLHKVLLDNLPTNVDLRFGTSVSEVTHGDDRAV
Mflv_2621|M.gilvum_PYR-GCK RRIVDALGEPLVVVRRSRLTDILREALPAGT-VRTGLAATRVSVGASSVR
Mvan_3960|M.vanbaalenii_PYR-1 QRIVHALGEPLVVVRRSALTEILLDALPPDT-VQTGLSATALSIAAATAR
MSMEG_4641|M.smegmatis_MC2_155 ERITRALGEPLVVIRRRVLTEILSGALAPGT-VVHGLEAHTVETCASGIR
MAV_4556|M.avium_104 T-------------------------LPKG--------------------
MAB_1601c|M.abscessus_ATCC_199 GKFTEAVGEPVAVTDRNQLLAILANRLTPGT-VRYGARVSNVRDGLHGTS
MMAR_4944|M.marinum_M QPLIEQVGQRPYPIARAELQQMLMEAYGIDE-IHFGMKMVEVANRDGAAT
TH_1880|M.thermoresistible__bu AELDTAHGYVLGIRQPVTDRLLAEWAAERGAVIRRGCAVVGLDQDAAGIT
MLBr_02276|M.leprae_Br4923 GPSFPSTSSAVPRLELDDRIRKVAENSGARMLLGTKAVSVYHDSSRRAAS
Mb0590c|M.bovis_AF2122/97 LTFERTAPRDFDLVIGADGLHSNVRRLVFGPERDFEHYLGCKVAACVVDG
Rv0575c|M.tuberculosis_H37Rv LTFERTAPRDFDLVIGADGLHSNVRRLVFGPERDFEHYLGCKVAACVVDG
MUL_2358|M.ulcerans_Agy99 VTLDDGARLDVDLLVGADGIHSTVRALAFGAEVQYLRYLGFHFAAFVFDA
Mflv_2621|M.gilvum_PYR-GCK ITLSDGEVRESDAVVGADGVNSVLARTLNGPLR--TRYVGYTAWRGIAAH
Mvan_3960|M.vanbaalenii_PYR-1 VTLSDGRTREADAVVGADGVNSMVARALNGPLP--SRYVGYTAWRGVAAY
MSMEG_4641|M.smegmatis_MC2_155 VTFSDGSVREASAVVGADGVDSVVARHLNGPLR--RRYAGYTAWRGIAAH
MAV_4556|M.avium_104 --------------------------------------------------
MAB_1601c|M.abscessus_ATCC_199 VELADGQSLTAAAVIGADGIGSLVAQYLNGPLA--FRYSGYTAWRGIADI
MMAR_4944|M.marinum_M ATFADGTIASADILIGADGANSITREYVLGGPVT-RRYAGYVNYNGLVEV
TH_1880|M.thermoresistible__bu VELADGSVQRARYLVGCDGGRSTVRKLAGIGFP--GEPARSETLLGEMSL
MLBr_02276|M.leprae_Br4923 VTLADGTEVGCRQLIIADGARSPLGRKLGRSWHQKTVYG--VAARGYLTT
Mb0590c|M.bovis_AF2122/97 YRPRDER--SYVLYNTVDRQLA-----RFALRGDRTMFLFVFRAEHDNPG
Rv0575c|M.tuberculosis_H37Rv YRPRDER--SYVLYNTVDRQLA-----RFALRGDRTMFLFVFRAEHDNPG
MUL_2358|M.ulcerans_Agy99 SDMDDTAGDHVVITDTVDRQLG-----LYALRNGQTAVFAAYRAADPEPP
Mflv_2621|M.gilvum_PYR-GCK PLDPELGG----ETLGPGTQVG-----HVPLGPDHTYWFATERTAEGGSA
Mvan_3960|M.vanbaalenii_PYR-1 RLDPALAG----ETMSAGTEVG-----HVPLGPDHTYWFATERTREGSRS
MSMEG_4641|M.smegmatis_MC2_155 PLDPELSG----ETMGAGVEVG-----HVPLGADHTYWFATERASQGQRS
MAV_4556|M.avium_104 --------------------------------------------------
MAB_1601c|M.abscessus_ATCC_199 SIPDELAG----LTVGPGIEFG-----HLPLSLGRTYWFAGERSLEAQRA
MMAR_4944|M.marinum_M DEAISPAN-EWTMYVGDGKRVS-----AMPVADDRFYFFFDVVEPEGLPF
TH_1880|M.thermoresistible__bu SADPATVAAVVAEVRKTQLRFG-----AMPLGDGVYRVIVPAEGVAGDRA
MLBr_02276|M.leprae_Br4923 PRSDDLWLTSHLELRSPDGAVLPGYGWIFPLGNGEVNIGVGALSTSKRPA
Mb0590c|M.bovis_AF2122/97 VA--PKDELRDQFGDVGWESRDILAALDDVEDLYFDVVSQIRMDRWSRGR
Rv0575c|M.tuberculosis_H37Rv VA--PKDELRDQFGDVGWESRDILAALDDVEDLYFDVVSQIRMDRWSRGR
MUL_2358|M.ulcerans_Agy99 PD--ARAALRDRFAGMGWLVPEVLERCPPSGDIYFGQVAQVEMPRWSTNR
Mflv_2621|M.gilvum_PYR-GCK PGG-EHAYLTAKVADWADPIPRLVATTDPGDLLRNDLYDRARAARWSDGR
Mvan_3960|M.vanbaalenii_PYR-1 AGG-EHAYLTAKLAGWADPIPALLASTDPADVLRNDLYDRAQPRDWSRGP
MSMEG_4641|M.smegmatis_MC2_155 PDG-ELTHLRRLFGSWAEPIPQLLAATDPADVLRNDLYDREPARCWARGP
MAV_4556|M.avium_104 --------------------------------------------------
MAB_1601c|M.abscessus_ATCC_199 PDG-EIEYLARKFGNWADPIPRLLRQSRESSVLRGDVYDRGRLRRVAGGR
MMAR_4944|M.marinum_M EKGTAREVLREQFAGWAPGVQALIDKLDPTTTNRVEILDLDPFHTWVKGR
TH_1880|M.thermoresistible__bu EPT--LDEFRRRLKAFAG---TDFGAHTPRWLSRFGDATR-LAERYRAGR
MLBr_02276|M.leprae_Br4923 DLA-----LRSLISYYTDLRRHEWGFSGQPRAVSSALLPMGGAVSGVAGP
Mb0590c|M.bovis_AF2122/97 -VLLIGDAAGCISLLGGEGTGLAITEAYVLAGELARAGG--DHRRAFDAY
Rv0575c|M.tuberculosis_H37Rv -VLLIGDAAGCISLLGGEGTGLAITEAYVLAGELARAGG--DHRRAFDAY
MUL_2358|M.ulcerans_Agy99 -VVLVGDACSAGSPLTGPGASLALAGAYVLSEQLRRTS---SVERALDFY
Mflv_2621|M.gilvum_PYR-GCK -AVLIGDAAHPMRPHLGQGGCQGIEDAAILARFLELAD---DVPTAFERF
Mvan_3960|M.vanbaalenii_PYR-1 -AVIVGDAAHPMRPHLGQGGCQGLEDAAILARFVGLSP---DLPAAFDRF
MSMEG_4641|M.smegmatis_MC2_155 -VVLAGDAAHPMRPHLGQGGCQGLEDAATLAALVAREP---DLPTAFARF
MAV_4556|M.avium_104 --------------------------------------------------
MAB_1601c|M.abscessus_ATCC_199 -VVLVGDAAHPMRPHLGQGGCQSLEDAAVLSVAISERS---SLPSAFREY
MMAR_4944|M.marinum_M -VAVLGDAAHNTTPDIGQGGCSAMEDAVALQWALKDNPT--DVAAALAAY
TH_1880|M.thermoresistible__bu -VFLAGDAAHVHPPAGGQGLNLGIQDAFNLGWKLAAAVGGWAPDGLLDTY
MLBr_02276|M.leprae_Br4923 NWMLIGDAAACVNPLNGEGIDYGLETGRLAAELLDLRDLSYVWPSLLQQH
Mb0590c|M.bovis_AF2122/97 EKRLRPFIEGKQASAAKFIWFFRHPN----PIRPVVSQRCDAHDELRPAG
Rv0575c|M.tuberculosis_H37Rv EKRLRPFIEGKQASAAKFIWFFATRTRFGLWFRNVAMRTMNFGPLATLFA
MUL_2358|M.ulcerans_Agy99 ERLWSPTVEEKQESARGLR----------LDAVAVVLTALDSPRRAALHL
Mflv_2621|M.gilvum_PYR-GCK EAFRKPRVGMLVREAQTLGRIVNVRPAVLSGLAGRATALIPERMVTAHLA
Mvan_3960|M.vanbaalenii_PYR-1 GAFRRRRVRPLVRESAAIGRIVNLRPAFLSGVASRASALIPEWAVASHLA
MSMEG_4641|M.smegmatis_MC2_155 ATLRRRRVTRMVRTSRMIGRIVNLRPAVLSAAASRATVLVPEAVLTRQLA
MAV_4556|M.avium_104 --------------------------------------------------
MAB_1601c|M.abscessus_ATCC_199 ARRRRSRTRAVVSRSRHIGNVTFARPAVVGGLLTRASARIPASVFWRQLS
MMAR_4944|M.marinum_M QASRTERAGDLVLRARKRCDVIHAKDPDTTAAWYEELRNEDGTNIIRGLV
TH_1880|M.thermoresistible__bu HAERHPVAADVLNNTRAQMELMSPEPG-PRAVRWLLAELMDFEEVNRYLT
MLBr_02276|M.leprae_Br4923 YGRGFSVARRLALLLTFKRCLPAIGPSAMRSTSLITIAVRVMANLVTDDD
Mb0590c|M.bovis_AF2122/97 DAVRRQRA------------------------------------------
Rv0575c|M.tuberculosis_H37Rv GSVRDDFELPDYTW------------------------------------
MUL_2358|M.ulcerans_Agy99 APADQPIR------------------------------------------
Mflv_2621|M.gilvum_PYR-GCK WIAAGSAFVLPTAADLAR--------------------------------
Mvan_3960|M.vanbaalenii_PYR-1 SIAAGSAFILPTAGDVGG--------------------------------
MSMEG_4641|M.smegmatis_MC2_155 SIAARSAFTPVD--------------------------------------
MAV_4556|M.avium_104 --------------------------------------------------
MAB_1601c|M.abscessus_ATCC_199 SIAGYEAGNLAVRP------------------------------------
MMAR_4944|M.marinum_M ANIMGGPLTPLS--------------------------------------
TH_1880|M.thermoresistible__bu EKITAIGVRYHTMGDGHGLLGRRMRDIPLTGGRLYARMHAGRGVLLDRTG
MLBr_02276|M.leprae_Br4923 ADWVARVWRSGGWASWLVDRRPPFS-------------------------
Mb0590c|M.bovis_AF2122/97 --------------------------------------------------
Rv0575c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2358|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2621|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3960|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4641|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4556|M.avium_104 --------------------------------------------------
MAB_1601c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4944|M.marinum_M --------------------------------------------------
TH_1880|M.thermoresistible__bu TLSVSGWADRVDHIVDPGAALDVPAVLLRPDGHVAWVGADHPGQQDLTAR
MLBr_02276|M.leprae_Br4923 --------------------------------------------------
Mb0590c|M.bovis_AF2122/97 ---------
Rv0575c|M.tuberculosis_H37Rv ---------
MUL_2358|M.ulcerans_Agy99 ---------
Mflv_2621|M.gilvum_PYR-GCK ---------
Mvan_3960|M.vanbaalenii_PYR-1 ---------
MSMEG_4641|M.smegmatis_MC2_155 ---------
MAV_4556|M.avium_104 ---------
MAB_1601c|M.abscessus_ATCC_199 ---------
MMAR_4944|M.marinum_M ---------
TH_1880|M.thermoresistible__bu LSRWFGAPA
MLBr_02276|M.leprae_Br4923 ---------