For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RVETSADVVVVGAGPAGSAAATWAARAGNDVIVIDSVCFPRDKACGDGLTPRAVAELKRLGLDDWLNTRI RHRGLRMSGFGGEVEVDWPGPSFPSTSSAVPRLELDDRIRKVAENSGARMLLGTKAVSVYHDSSRRAASV TLADGTEVGCRQLIIADGARSPLGRKLGRSWHQKTVYGVAARGYLTTPRSDDLWLTSHLELRSPDGAVLP GYGWIFPLGNGEVNIGVGALSTSKRPADLALRSLISYYTDLRRHEWGFSGQPRAVSSALLPMGGAVSGVA GPNWMLIGDAAACVNPLNGEGIDYGLETGRLAAELLDLRDLSYVWPSLLQQHYGRGFSVARRLALLLTFK RCLPAIGPSAMRSTSLITIAVRVMANLVTDDDADWVARVWRSGGWASWLVDRRPPFS*
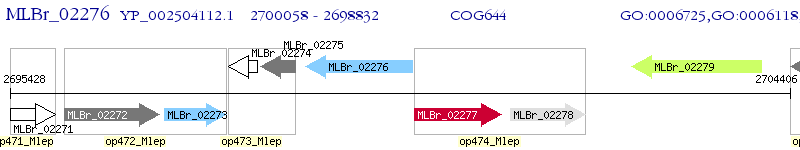
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_02276 | - | - | 100% (408) | putative FAD-linked oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0576c | - | 0.0 | 84.31% (408) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_5254 | - | 1e-170 | 72.73% (407) | geranylgeranyl reductase |
| M. tuberculosis H37Rv | Rv0561c | - | 0.0 | 84.31% (408) | oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3929 | - | 1e-157 | 65.53% (409) | oxidoreductase |
| M. marinum M | MMAR_0910 | - | 0.0 | 84.04% (401) | FAD-linked oxidoreductase |
| M. avium 104 | MAV_4582 | - | 0.0 | 79.85% (407) | FAD dependent oxidoreductase, putative |
| M. smegmatis MC2 155 | MSMEG_1132 | - | 1e-180 | 74.57% (409) | FAD binding domain-containing protein |
| M. thermoresistible (build 8) | TH_0132 | - | 1e-138 | 73.91% (322) | POSSIBLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_0663 | - | 0.0 | 83.29% (401) | FAD-linked oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_0991 | - | 1e-171 | 73.76% (404) | geranylgeranyl reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0576c|M.bovis_AF2122/97 ----------MSVDDSADVVVVGAGPAGSAAAAWAARAGRDVLVIDTATF
Rv0561c|M.tuberculosis_H37Rv ----------MSVDDSADVVVVGAGPAGSAAAAWAARAGRDVLVIDTATF
MMAR_0910|M.marinum_M ------METRVPRALGADLVVVGAGPAGSAAAAWAARAGRDVLVIDSAGF
MUL_0663|M.ulcerans_Agy99 ------METRVPRALGADLVVVGAGPAGSAAAAWAARAGRDVLVIDSAGF
MLBr_02276|M.leprae_Br4923 ----------MRVETSADVVVVGAGPAGSAAATWAARAGNDVIVIDSVCF
MAV_4582|M.avium_104 -------MCMTTAATRADLVVVGAGPAGSAAAAWAARAGHDVLVVDAARF
Mflv_5254|M.gilvum_PYR-GCK MDTPA--VGAIRGSSGADVVVIGAGPAGSSAAAWAARSGRDVLVIDSAQF
Mvan_0991|M.vanbaalenii_PYR-1 MTSAAGRSAATGDSTAVDVVVVGAGPAGSSAAAWAARAGRDVLVIDSAQF
MSMEG_1132|M.smegmatis_MC2_155 ------------MNTRADVVVVGAGPAGSAAAAWAARAGRDVVVVDAAQF
TH_0132|M.thermoresistible__bu --------------------------------------------------
MAB_3929|M.abscessus_ATCC_1997 ------------MQTSADLVVVGAGPAGSAAAAWAARSGREVVLVDSAVF
Mb0576c|M.bovis_AF2122/97 PRDKPCGDGLTPRAVAELHQLGLGKWLADHIRHRGLRMSGFGGEVEVDWP
Rv0561c|M.tuberculosis_H37Rv PRDKPCGDGLTPRAVAELHQLGLGKWLADHIRHRGLRMSGFGGEVEVDWP
MMAR_0910|M.marinum_M PRDKACGDGLTPRAIAELQQLGLGEWLAGRIQHRGLRMSGFGGEVEIGWP
MUL_0663|M.ulcerans_Agy99 PRDKACGDGLTPRAIAELQQLGLGEWLAGRIQHRGLRMSGFGGEVEIGWP
MLBr_02276|M.leprae_Br4923 PRDKACGDGLTPRAVAELKRLGLDDWLNTRIRHRGLRMSGFGGEVEVDWP
MAV_4582|M.avium_104 PRDKACGDGLTPRAVAECERLGLGGWLDSRIRHRGLRMSGFGAQVQVDWP
Mflv_5254|M.gilvum_PYR-GCK PRDKACGDGLTPRAVAELDRLGLRSWLDGRIAHRGLRMSGFGSDVEVPWP
Mvan_0991|M.vanbaalenii_PYR-1 PRDKACGDGLTPRAIAELRRLGLQSWLDGRIAHRGLRMSGFGADVEVAWP
MSMEG_1132|M.smegmatis_MC2_155 PRDKACGDGLTPRAVAELQRLGMASWLDTRIRHHGLRMSGFGADVEIPWP
TH_0132|M.thermoresistible__bu ----------------------------------------------VPWP
MAB_3929|M.abscessus_ATCC_1997 PRDKTCGDGLTPRAVEQLELLGLRPWLDEHIRYRGLRLHGFGGNLEVPWP
: **
Mb0576c|M.bovis_AF2122/97 GPSFPSYGSAVARLELDDRIRKVAEDTGARMLLGAKAVAVHHDSSRRVVS
Rv0561c|M.tuberculosis_H37Rv GPSFPSYGSAVARLELDDRIRKVAEDTGARMLLGAKAVAVHHDSSRRVVS
MMAR_0910|M.marinum_M GPSFPAYSSAVARLELDDRIRKVAEESGARMLLGTKAVAAHHDSSRRVVS
MUL_0663|M.ulcerans_Agy99 GPSFPAYSSAVARLELDDRIRKVAEESGARMLLGTKAVAAHHDSSRRVVS
MLBr_02276|M.leprae_Br4923 GPSFPSTSSAVPRLELDDRIRKVAENSGARMLLGTKAVSVYHDSSRRAAS
MAV_4582|M.avium_104 GPSFPSTGSAVARTELDDRIRRVAEDSGARMLLGTKAVGVQRDSSAAVTS
Mflv_5254|M.gilvum_PYR-GCK GPSFPPTSSAVPRTELDERIRLVAVDDGAKMKLGVKAVDVERDSTGRVTG
Mvan_0991|M.vanbaalenii_PYR-1 GPSFPSISSAVPRTELDERIRLVAVDDGAKMKLGVKAVSVERGSGGRVTG
MSMEG_1132|M.smegmatis_MC2_155 GPSFPATSSAVPRTELDDRIRSVAADDGAKMMLGTKVVDVTHDSSGRVDA
TH_0132|M.thermoresistible__bu GPSFPSTGSAGPRTELDDRIRMVAVDDGAKMMLGVKAAGVTHDSSGRVAS
MAB_3929|M.abscessus_ATCC_1997 GTSFGGTGSAVRRTELDDKIRQVAVDSGAKMAESTKAVDVERDSAGRITS
*.** .** * ***::** ** : **:* ..*.. . :.* .
Mb0576c|M.bovis_AF2122/97 LTLADGT---EVGCRQLIVADGARSPLGRKLGRRWHRETVYGVAVRGYLS
Rv0561c|M.tuberculosis_H37Rv LTLADGT---EVGCRQLIVADGARSPLGRKLGRRWHRETVYGVAVRGYLS
MMAR_0910|M.marinum_M LTLADRT---EVACRQLIVADGARSSLGRKLGRRWHQETVYGVAARGYLT
MUL_0663|M.ulcerans_Agy99 LTLADRT---EVACRQLIVADGARSSLGRKLGRRWHQETVYGVAARGYLT
MLBr_02276|M.leprae_Br4923 VTLADGT---EVGCRQLIIADGARSPLGRKLGRSWHQKTVYGVAARGYLT
MAV_4582|M.avium_104 LMLADGT---GVQCRQLIVADGARSSLGRTLGRRWHQETVYGVAARGYLS
Mflv_5254|M.gilvum_PYR-GCK VVLDSGE---LVRCSSLIVADGARSTLGRTLGRQWHKETVYGVAIRGYIG
Mvan_0991|M.vanbaalenii_PYR-1 VVLDTGE---VVRCRDLIVADGARSTLGRALGREWHKETVYGVAIRGYIA
MSMEG_1132|M.smegmatis_MC2_155 VVLDDGN---TVGCAQLIVADGARSTLGRVLGRTWHRETVYGVAIRGYIA
TH_0132|M.thermoresistible__bu VQLDNGT---EIRCTELIVADGARSTLGRALGRQWHQQTVYGIAIRAYAE
MAB_3929|M.abscessus_ATCC_1997 VRVLDDNGPGEIACKAVIVADGVRSTLGKILGREWHKDTVYAVAGRAYIE
: : : * :*:***.**.**: *** **:.***.:* *.*
Mb0576c|M.bovis_AF2122/97 TAYSDDPWLTSHLELRSPDGA---VLPGYGWIFPLGNGEVNIGVGALSTS
Rv0561c|M.tuberculosis_H37Rv TAYSDDPWLTSHLELRSPDGA---VLPGYGWIFPLGNGEVNIGVGALSTS
MMAR_0910|M.marinum_M TARSEDPWLTSHLELRSPDGA---VLPGYGWIFPLGNGEVNIGVGALSTT
MUL_0663|M.ulcerans_Agy99 TARSEDPWLTSHLELRSPDGA---VLPGYGWIFPLGNGEVNIGVGGLSTT
MLBr_02276|M.leprae_Br4923 TPRSDDLWLTSHLELRSPDGA---VLPGYGWIFPLGNGEVNIGVGALSTS
MAV_4582|M.avium_104 TPRADDPWLTSHLELRGPDGARDKVLPGYGWIFPLGNGEVNIGVGALSTS
Mflv_5254|M.gilvum_PYR-GCK TPRSDEEWITSHLELRSPAGE---VLPGYGWIFPLGNGEVNIGVGALATS
Mvan_0991|M.vanbaalenii_PYR-1 TPRSDEPWITSHLELRSPAGE---VLPGYGWIFPLGNGEVNIGVGALATA
MSMEG_1132|M.smegmatis_MC2_155 SPRASEPWITSHLELRSPEGK---VLPGYGWMFPLGNGEVNIGVGALATA
TH_0132|M.thermoresistible__bu SPRHDEPWITSHLELRSAEGE---LLPGYGWIFPLGNGEVNIGVGALSTA
MAB_3929|M.abscessus_ATCC_1997 SARGDEEWMTSHLELRSPEGD---VLPGYGWIFPLGGGQINIGVGALATS
:. .: *:*******.. * :******:****.*::*****.*:*:
Mb0576c|M.bovis_AF2122/97 RRPADLALRPLISYYTDLRRDEWGFTGQPRAVSSALLPMGGAVSGVAGSN
Rv0561c|M.tuberculosis_H37Rv RRPADLALRPLISYYTDLRRDEWGFTGQPRAVSSALLPMGGAVSGVAGSN
MMAR_0910|M.marinum_M KRPADLALRPLMSYYTDLRRDEWGFTGQPRAVASALLPMGGAVSGVAGPN
MUL_0663|M.ulcerans_Agy99 KRPADLALRPLMSYYTDLRRDEWGFTGQPRAVASALLPMGGAVSGVAGPN
MLBr_02276|M.leprae_Br4923 KRPADLALRSLISYYTDLRRHEWGFSGQPRAVSSALLPMGGAVSGVAGPN
MAV_4582|M.avium_104 RRPAELALRPLIDHYTDLRRDEWGFVGRPRAVASALLPMGGAVSGVAGPN
Mflv_5254|M.gilvum_PYR-GCK KRPADAALRPLLNYYTGLRRAEWGFEGDPRAPLSALLPMGGAVSRVAGPN
Mvan_0991|M.vanbaalenii_PYR-1 KRPADAALRPLMNYYAGLRRDEWGFSGDPRAGLSALLPMGGAVSGVAGPN
MSMEG_1132|M.smegmatis_MC2_155 KRPADAALRPLMSYYADLRREEWGLVGEPRAGLSALLPMGGAVSGVAGPN
TH_0132|M.thermoresistible__bu KRPADAALRPLLSFYTGLRRDEWGFPGPPRAALSALLPMGGAVSGVAGPN
MAB_3929|M.abscessus_ATCC_1997 KRPADAALRPLIEYYTELRREDFGLTGRLQSIASALLPMGGAVSGVAGPN
:***: ***.*:..*: *** ::*: * :: *********** ***.*
Mb0576c|M.bovis_AF2122/97 WMLIGDAAACVNPLNGEGIDYGLETGRLAAELLDSR---DLARLWPSLLA
Rv0561c|M.tuberculosis_H37Rv WMLIGDAAACVNPLNGEGIDYGLETGRLAAELLDSR---DLARLWPSLLA
MMAR_0910|M.marinum_M WMLIGDAAACVNPLNGEGIDYGLETGRLAAELLDSP---DLSQLWPSLLQ
MUL_0663|M.ulcerans_Agy99 WMLIGDAAACVNPLNGEGIDYGLDTGRLAAELLDSP---DLSQLWPSLLQ
MLBr_02276|M.leprae_Br4923 WMLIGDAAACVNPLNGEGIDYGLETGRLAAELLDLR---DLSYVWPSLLQ
MAV_4582|M.avium_104 WMLIGDAAACVNPLNGEGIDYGMETGRLAAGLLGCP---DLSQEWPALLR
Mflv_5254|M.gilvum_PYR-GCK WMLIGDAAACVNPLNGEGIDYGLETGRLAVSLLGTG---SFEASWPALLH
Mvan_0991|M.vanbaalenii_PYR-1 WMLIGDAAACVNPLNGEGIDYGLETGRLAASLLGSG---SYSAAWPALLH
MSMEG_1132|M.smegmatis_MC2_155 WMLIGDAAACVNPLNGEGIDYGLETGRLAAELMTSGGVTDYSSAWPTLLQ
TH_0132|M.thermoresistible__bu WMLIGDAAACVNPLNGEGIDYGLETGRFAAGLLGSG---DLSQAWPALLQ
MAB_3929|M.abscessus_ATCC_1997 WMLIGDAAACVNPLNGEGIDYGLETGHLAAEVLDAG---DYSQLWPQLLR
**********************::**::*. :: . ** **
Mb0576c|M.bovis_AF2122/97 DRYGRGFSVARRLALLLTFPRFLPTTGPITMRSTALMNIAVRVMSNLVTD
Rv0561c|M.tuberculosis_H37Rv DRYGRGFSVARRLALLLTFPRFLPTTGPITMRSTALMNIAVRVMSNLVTD
MMAR_0910|M.marinum_M AHYGRGFSVARRLALLLTYQRFLPTTGPVAMRSTALMKVAVRVMANLVTE
MUL_0663|M.ulcerans_Agy99 AHYGRGFSVARRLALLLTYQRFLPTTGPVAMRSTALMKVAMRVMANLVTE
MLBr_02276|M.leprae_Br4923 QHYGRGFSVARRLALLLTFKRCLPAIGPSAMRSTSLITIAVRVMANLVTD
MAV_4582|M.avium_104 EHYARGFSVARRLALLLTYQRFLPATGPVAMRSTTLMRIAVRVMANLVTD
Mflv_5254|M.gilvum_PYR-GCK EHYARGFSVARRLALLLTFPRFLPVAGPVAMRSSTLMKIAVRVMGNFVTD
Mvan_0991|M.vanbaalenii_PYR-1 GHYARGFSIARRLALLLTFPRFLPAAGPLAMRSSALMKTAVRVMGNFVTD
MSMEG_1132|M.smegmatis_MC2_155 EHYARGFSVARRLALLLTLPRFLQVTGPVAMRSATLMTIAVRVMGNLVTD
TH_0132|M.thermoresistible__bu RHYGHGFSVARRLASLLTRPRFLPLTGPVAMRSTKLMNIAVRVMGNLVTD
MAB_3929|M.abscessus_ATCC_1997 DRYGRSFSIARRLAALLTLPRFLPVMGPVGMHSAVLMRIAVRLMGNLVTD
:*.:.**:***** *** * * ** *:*: *: *:*:*.*:**:
Mb0576c|M.bovis_AF2122/97 DDRDWVARVWRGGGQLSRLVDRRPPFS
Rv0561c|M.tuberculosis_H37Rv DDRDWVARVWRGGGQLSRLVDRRPPFS
MMAR_0910|M.marinum_M EDSDWVARLWRGGGRASRLIDRRPPFH
MUL_0663|M.ulcerans_Agy99 EDSDWVARLWRGGGRASRLIDRRPPFH
MLBr_02276|M.leprae_Br4923 DDADWVARVWRSGGWASWLVDRRPPFS
MAV_4582|M.avium_104 DDADWVARAWRGAGRLSRLIDRRAPFS
Mflv_5254|M.gilvum_PYR-GCK EDEDAVARVWRGAGRLSGRIDERRPFV
Mvan_0991|M.vanbaalenii_PYR-1 EDEDAVARVWRGAGTLSRRMDARRPFT
MSMEG_1132|M.smegmatis_MC2_155 EDADWIARVWRTAGLASRRIDQRVPFS
TH_0132|M.thermoresistible__bu EDSDWVARVWRTAGAGSRRLDERPPFS
MAB_3929|M.abscessus_ATCC_1997 DDQDMIARAWRVAGGGSRRLDSRPPFS
:* * :** ** .* * :* * **