For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNGRDPDAAHEAAQRIPGALAHPGSPADPAVADALIDICVREFGRIDILINCAGTAEPRGSSILNVTSAQ FRELLDAHLGTTFETCRAAAPVMVAQGGGAIVNTGSFAFLGDYGGTGYPAGKGAVNSLTLAIAAELAEHG VRANVVCPGAKTRLSTGPEYESHITELNRRGLLDDVSFQGALDAAPPEYVAPTYAYLASDLATQVTGQIF IAAGGFVGRFERQTPSLVAYRGHDDEPPWTVEEIAAQVR*
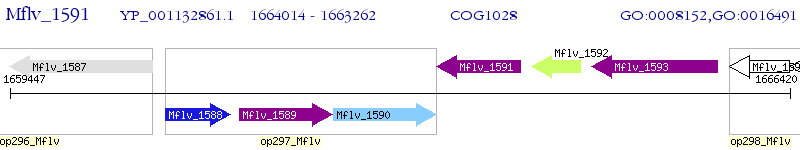
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1591 | - | - | 100% (250) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_1790 | - | 1e-24 | 32.94% (252) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_2001 | - | 2e-21 | 34.25% (219) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 1e-20 | 31.19% (218) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv2857c | - | 1e-20 | 31.19% (218) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 1e-11 | 27.62% (210) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_1210c | - | 4e-81 | 59.84% (249) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_2633 | - | 7e-88 | 60.47% (253) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_2962 | - | 4e-89 | 62.85% (253) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_5869 | - | 1e-111 | 79.59% (245) | putative short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1252 | - | 1e-111 | 79.18% (245) | putative short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_3108 | - | 2e-85 | 59.29% (253) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_5166 | - | 1e-120 | 83.53% (249) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Mb2882c|M.bovis_AF2122/97 ---MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVVGDVD--VE
Rv2857c|M.tuberculosis_H37Rv ---MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVVGDVD--VE
Mflv_1591|M.gilvum_PYR-GCK ----------------------------------------MVNGRD--PD
Mvan_5166|M.vanbaalenii_PYR-1 ----------------MVVGGTRGVGLAVAELLAALGAGVVVNGRD--EV
MSMEG_5869|M.smegmatis_MC2_155 ----------------MVVGGSRGIGLAVAGLLAAQGAGVVVNGRD--AD
TH_1252|M.thermoresistible__bu ---------------------------------------VVVNGRD--PE
MMAR_2633|M.marinum_M MTDRD----LLAGRGVVVVGGSRGIGRAVAELLARLGAGVVISGRE--PD
MUL_3108|M.ulcerans_Agy99 MTDRD----LLAGRGVVVVGGSRGIGRAVAELLARLGAGVVISGRE--PD
MAB_1210c|M.abscessus_ATCC_199 MPN------------CVVVGGSRGIGRAVALLLGELGTNVVVNGRH--AD
MAV_2962|M.avium_104 MAAVSGAAALLSGRGAVVSGGSRGIGRAVAELLAGLGAGVVVNGRD--PQ
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVESAAE
: . .
Mb2882c|M.bovis_AF2122/97 AGGAAADELSG-----LFVPTDVCDEDAVNGLFDGAAETYG-RIDIAFNN
Rv2857c|M.tuberculosis_H37Rv AGGAAADELSG-----LFVPTDVCDEDAVNGLFDGAAETYG-RIDIAFNN
Mflv_1591|M.gilvum_PYR-GCK AAHEAAQRIPG----ALAHPGSPADPAVADALIDICVREFG-RIDILINC
Mvan_5166|M.vanbaalenii_PYR-1 AAGQAAERIPR----AIGFSGSPSDPDVADALIDTCVREFG-RIDILVNC
MSMEG_5869|M.smegmatis_MC2_155 AAEAAAKRIS-----GVAHAGSPADPAVADALVDTCVTEFG-RIDILVNC
TH_1252|M.thermoresistible__bu VVTEAAQRIS-----GVGHAGSPADPAVADALVDTCVQNFG-SIDILINC
MMAR_2633|M.marinum_M AVRDATEAVISAGGRAAGIAGAADHEDTANALVDRCMTSFG-RLDALINC
MUL_3108|M.ulcerans_Agy99 AVRDATEAVISAGGRAAGIAGAADHEDTANALVDRCMTSFG-RLDALINC
MAB_1210c|M.abscessus_ATCC_199 VVDDTVGAITSSGGTAVGFVGAPDNERTATDLIDTCRTQYG-GIDALINC
MAV_2962|M.avium_104 AVRETVAAITAAGGRATAVVGAADDERIARSLVDECIGAFG-RLDALINC
MLBr_02565|M.leprae_Br4923 ALDETANRVGG-----TTLSLDVTADDAVDKITEHLHDYQGGHADILVNN
. :. : . : : * * .*
Mb2882c|M.bovis_AF2122/97 AGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLAGKGSIV
Rv2857c|M.tuberculosis_H37Rv AGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLAGKGSIV
Mflv_1591|M.gilvum_PYR-GCK AGTAEPRGSSILNVTSAQFRELLDAHLGTTFETCRAAAPVMVAQGGGAIV
Mvan_5166|M.vanbaalenii_PYR-1 AGTAEPVGSSILNVTPAQFQELIDAHLGTAFQTCRAAAPLMARQGSGAIV
MSMEG_5869|M.smegmatis_MC2_155 AGTAEPHGSSILDITSAQFRELLDAHLGTVFETCRAAAPKMVAHGGGAIV
TH_1252|M.thermoresistible__bu AGTAEPPGSSILNISSGQFRELLDAHLGTVFETCRAAAPRMVAQGGGAIV
MMAR_2633|M.marinum_M AGIAEPPGSSILNISPAQFDGLLRAHLGTVFHTCRVAAPVLAAQGHGSII
MUL_3108|M.ulcerans_Agy99 AGIAEPPGSSILNISPAQSDGLLRAHLGTVFHACRVAAPVLAAQGHGSII
MAB_1210c|M.abscessus_ATCC_199 AGIPEPTGSSILTITSGDFHRLIDAHLGTVFHTCRVVAPLMVQQRHGAII
MAV_2962|M.avium_104 AGIAEPAGSSILNITADEFDHLIGAHLGTAFHTCRAAAPVMVEQRHGSIV
MLBr_02565|M.leprae_Br4923 AGITR--DKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSISKGGRVI
** . .. : : .:* : . * ::
Mb2882c|M.bovis_AF2122/97 NTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVNALCPGP
Rv2857c|M.tuberculosis_H37Rv NTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVNALCPGP
Mflv_1591|M.gilvum_PYR-GCK NTGSFAFLG--DYGGTGYPAGKGAVNSLTLAIAAELAEHGVRANVVCPGA
Mvan_5166|M.vanbaalenii_PYR-1 NTSSFAFLG--DYGGTGYAAGKGAVNSLTLAIAAELKEHGIRANVVCPGA
MSMEG_5869|M.smegmatis_MC2_155 NTGSFAYLG--DYGGTGYPAGKGGVNALTMAIAAELREHKVRANVVCPGA
TH_1252|M.thermoresistible__bu NTSSFAFLG--DYGGTGYPAGKGGVNGLTLAIAAELKEHSVRANVVCPGA
MMAR_2633|M.marinum_M NTGSVAFLG--DYGGSGYPAGKAAVNGLTLAISAELKARGVRANVICPGA
MUL_3108|M.ulcerans_Agy99 NTGSVAFLG--DYGGSGYPAGKAAVNGLTLAISAELKARGVRANVICPGA
MAB_1210c|M.abscessus_ATCC_199 NTGSTASLG--IYGGTGYPAGKAAVNGLTLAIAAELKRSGVRANVICPGA
MAV_2962|M.avium_104 NTSSVAFLG--DYGGTGYPAGKGAVNALTMAIAAELKAYGVRANVVCPGA
MLBr_02565|M.leprae_Br4923 VLSSMAGIAG-NRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPGF
.* . : . .*.:.*..* .:: :. : : *.:.**
Mb2882c|M.bovis_AF2122/97 VNTPLLQELFAKNPERAARR-----MVHVPLGRFAEPDEIAAAVAFLASD
Rv2857c|M.tuberculosis_H37Rv VNTPLLQELFAKNPERAARR-----MVHVPLGRFAEPDEIAAAVAFLASD
Mflv_1591|M.gilvum_PYR-GCK KTRLSTGPEYESHITELNRRGLLDDVSFQGALDAAPPEYVAPTYAYLASD
Mvan_5166|M.vanbaalenii_PYR-1 KTRLSTGSDYESHIAELNRRGMLDDVSYQGALDAAPPEYAAPTYAYLVSD
MSMEG_5869|M.smegmatis_MC2_155 KTRLSTGPEYERHIAELNRRGLLDDVSAQGALDAAAPEYAAATYAYLVSD
TH_1252|M.thermoresistible__bu RTRLSTGPEYEAHIAELHRRGLLDEVSMQGALDAAPPEYVAPTYAYLVSD
MMAR_2633|M.marinum_M RTRLSSGPDYEAHINELHRRGLLDEMTRQASLDAPPADFVAPLYAYLASD
MUL_3108|M.ulcerans_Agy99 RTRLSSGPNYEAHINELHRRGLLDEMTRQASLDAPPADFVAPLYAYLASD
MAB_1210c|M.abscessus_ATCC_199 RTRLSEGPEYTAHLHDLHTRGLLDDMTLQASLVPAPPACVAPLYAYLAGD
MAV_2962|M.avium_104 RTRLSTGADYERHIEDLHRRGLLDDMTRRASLDSAPPVFVAPVYGYLVSD
MLBr_02565|M.leprae_Br4923 IETQMT-AAIPLATREVGRR-------LNSLLQGGLPVDVAETIAYFAIP
* . * .::.
Mb2882c|M.bovis_AF2122/97 DASFITASTFLVDGGISSAYVTPL--------------------------
Rv2857c|M.tuberculosis_H37Rv DASFITASTFLVDGGISSAYVTPL--------------------------
Mflv_1591|M.gilvum_PYR-GCK LATQVTGQIFIAAGGFVGRFERQTPSLVAYRGHDDEPPWTVEEIAAQVR-
Mvan_5166|M.vanbaalenii_PYR-1 LANHITGQIFIAAGGFVGRFDRQTPALVAYRGHHDEPPWTVEEIAAQVG-
MSMEG_5869|M.smegmatis_MC2_155 AARDVTGRIFIAAGGFVGEFARPTPGFLGFRDHHTSPPWTADELHQMISG
TH_1252|M.thermoresistible__bu LAKDITGRIFIAAGGFLGEFARPTPDFLGFRDANDSPPFSVEEIHDMINR
MMAR_2633|M.marinum_M LASGITGQIFVASGGFVGTFDRPTPRVLGWRDHRADPPWPVEDLHTMINA
MUL_3108|M.ulcerans_Agy99 LASGITGQIFVASGGSVGTFDRPTPRVLGWRDHRADPPWPVEDLHTMINA
MAB_1210c|M.abscessus_ATCC_199 LSARVTGQIFLAAGGFIGRFKRQTPVPLGYRDHALDAPWSMEEIDDLMGR
MAV_2962|M.avium_104 LARDVTGQILVAAGGFVGSFDRQTPRLLGYRDHHRAGPWSIEEVHTMIGA
MLBr_02565|M.leprae_Br4923 ASNAVTGNVIRVCGQAMLGA------------------------------
: :*. : . *
Mb2882c|M.bovis_AF2122/97 ------
Rv2857c|M.tuberculosis_H37Rv ------
Mflv_1591|M.gilvum_PYR-GCK ------
Mvan_5166|M.vanbaalenii_PYR-1 ------
MSMEG_5869|M.smegmatis_MC2_155 G-----
TH_1252|M.thermoresistible__bu ------
MMAR_2633|M.marinum_M RLLDTD
MUL_3108|M.ulcerans_Agy99 RLLDTD
MAB_1210c|M.abscessus_ATCC_199 R-----
MAV_2962|M.avium_104 AATP--
MLBr_02565|M.leprae_Br4923 ------