For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDRDLLAGRGVVVVGGSRGIGRAVAELLARLGAGVVISGREPDAVRDATEAVISAGGRAAGIAGAADHE DTANALVDRCMTSFGRLDALINCAGIAEPPGSSILNISPAQFDGLLRAHLGTVFHTCRVAAPVLAAQGHG SIINTGSVAFLGDYGGSGYPAGKAAVNGLTLAISAELKARGVRANVICPGARTRLSSGPDYEAHINELHR RGLLDEMTRQASLDAPPADFVAPLYAYLASDLASGITGQIFVASGGFVGTFDRPTPRVLGWRDHRADPPW PVEDLHTMINARLLDTD
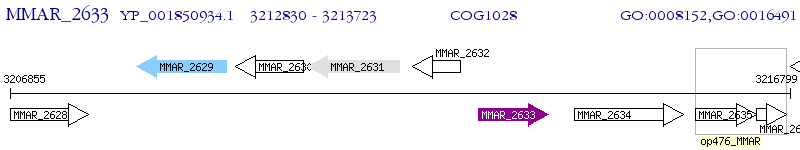
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2633 | - | - | 100% (297) | short-chain type dehydrogenase/reductase |
| M. marinum M | MMAR_0629 | - | 1e-24 | 34.25% (254) | dehydrogenase |
| M. marinum M | MMAR_5036 | - | 1e-24 | 34.07% (270) | short-chain type dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2788c | fabG | 3e-27 | 34.73% (262) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1591 | - | 8e-88 | 60.47% (253) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2766c | fabG | 4e-27 | 34.73% (262) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_00862 | ephD | 2e-19 | 31.56% (225) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1210c | - | 1e-101 | 64.29% (280) | putative short chain dehydrogenase/reductase |
| M. avium 104 | MAV_2962 | - | 1e-118 | 70.98% (286) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_5869 | - | 4e-94 | 60.71% (280) | putative short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1252 | - | 3e-91 | 63.53% (255) | putative short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_3108 | - | 1e-167 | 98.65% (297) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_5166 | - | 3e-98 | 61.01% (277) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mb2788c|M.bovis_AF2122/97 --------------------MTSLDLTGRTAIITGASRGIGLAIAQQLAA
Rv2766c|M.tuberculosis_H37Rv --------------------MTSLDLTGRTAIITGASRGIGLAIAQQLAA
MMAR_2633|M.marinum_M -------------------MTDRDLLAGRGVVVVGGSRGIGRAVAELLAR
MUL_3108|M.ulcerans_Agy99 -------------------MTDRDLLAGRGVVVVGGSRGIGRAVAELLAR
MAB_1210c|M.abscessus_ATCC_199 ---------------------------MPNCVVVGGSRGIGRAVALLLGE
MAV_2962|M.avium_104 ---------------MAAVSGAAALLSGRGAVVSGGSRGIGRAVAELLAG
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 -------------------------------MVVGGTRGVGLAVAELLAA
MSMEG_5869|M.smegmatis_MC2_155 -------------------------------MVVGGSRGIGLAVAGLLAA
TH_1252|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
Mb2788c|M.bovis_AF2122/97 AGAHVVLTARRQEAADEAAAQVGDR---ALGVGAHAVDEDAARRCVDLTL
Rv2766c|M.tuberculosis_H37Rv AGAHVVLTARRQEAADEAAAQVGDR---ALGVGAHAVDEDAARRCVDLTL
MMAR_2633|M.marinum_M LGAGVVISGREPDAVRDATEAVISAGGRAAGIAGAADHEDTANALVDRCM
MUL_3108|M.ulcerans_Agy99 LGAGVVISGREPDAVRDATEAVISAGGRAAGIAGAADHEDTANALVDRCM
MAB_1210c|M.abscessus_ATCC_199 LGTNVVVNGRHADVVDDTVGAITSSGGTAVGFVGAPDNERTATDLIDTCR
MAV_2962|M.avium_104 LGAGVVVNGRDPQAVRETVAAITAAGGRATAVVGAADDERIARSLVDECI
Mflv_1591|M.gilvum_PYR-GCK -----MVNGRDPDAAHEAAQRIPG----ALAHPGSPADPAVADALIDICV
Mvan_5166|M.vanbaalenii_PYR-1 LGAGVVVNGRDEVAAGQAAERIPR----AIGFSGSPSDPDVADALIDTCV
MSMEG_5869|M.smegmatis_MC2_155 QGAGVVVNGRDADAAEAAAKRIS-----GVAHAGSPADPAVADALVDTCV
TH_1252|M.thermoresistible__bu ----VVVNGRDPEVVTEAAQRIS-----GVGHAGSPADPAVADALVDTCV
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
::. .. :. : . . . . :
Mb2788c|M.bovis_AF2122/97 ERFGSVDILINNAGTNPAYG-PLLEQDHARFAKIFDVNLWAPLMWTSLVV
Rv2766c|M.tuberculosis_H37Rv ERFGSVDILINNAGTNPAYG-PLLEQDHARFAKIFDVNLWAPLMWTSLVV
MMAR_2633|M.marinum_M TSFGRLDALINCAGIAEPPGSSILNISPAQFDGLLRAHLGTVFHTCRVAA
MUL_3108|M.ulcerans_Agy99 TSFGRLDALINCAGIAEPPGSSILNISPAQSDGLLRAHLGTVFHACRVAA
MAB_1210c|M.abscessus_ATCC_199 TQYGGIDALINCAGIPEPTGSSILTITSGDFHRLIDAHLGTVFHTCRVVA
MAV_2962|M.avium_104 GAFGRLDALINCAGIAEPAGSSILNITADEFDHLIGAHLGTAFHTCRAAA
Mflv_1591|M.gilvum_PYR-GCK REFGRIDILINCAGTAEPRGSSILNVTSAQFRELLDAHLGTTFETCRAAA
Mvan_5166|M.vanbaalenii_PYR-1 REFGRIDILVNCAGTAEPVGSSILNVTPAQFQELIDAHLGTAFQTCRAAA
MSMEG_5869|M.smegmatis_MC2_155 TEFGRIDILVNCAGTAEPHGSSILDITSAQFRELLDAHLGTVFETCRAAA
TH_1252|M.thermoresistible__bu QNFGSIDILINCAGTAEPPGSSILNISSGQFRELLDAHLGTVFETCRAAA
MLBr_00862|M.leprae_Br4923 AKHGVPDIVVNNAGIG--QAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFG
.* * ::* ** . :* :: .:* .
Mb2788c|M.bovis_AF2122/97 TAWMGEHGG-AVVNTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSP
Rv2766c|M.tuberculosis_H37Rv TAWMGEHGG-AVVNTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSP
MMAR_2633|M.marinum_M PVLAAQGHG-SIINTGSVAFLGDYGGSG-YPAGKAAVNGLTLAISAELKA
MUL_3108|M.ulcerans_Agy99 PVLAAQGHG-SIINTGSVAFLGDYGGSG-YPAGKAAVNGLTLAISAELKA
MAB_1210c|M.abscessus_ATCC_199 PLMVQQRHG-AIINTGSTASLGIYGGTG-YPAGKAAVNGLTLAIAAELKR
MAV_2962|M.avium_104 PVMVEQRHG-SIVNTSSVAFLGDYGGTG-YPAGKGAVNALTMAIAAELKA
Mflv_1591|M.gilvum_PYR-GCK PVMVAQGGG-AIVNTGSFAFLGDYGGTG-YPAGKGAVNSLTLAIAAELAE
Mvan_5166|M.vanbaalenii_PYR-1 PLMARQGSG-AIVNTSSFAFLGDYGGTG-YAAGKGAVNSLTLAIAAELKE
MSMEG_5869|M.smegmatis_MC2_155 PKMVAHGGG-AIVNTGSFAYLGDYGGTG-YPAGKGGVNALTMAIAAELRE
TH_1252|M.thermoresistible__bu PRMVAQGGG-AIVNTSSFAFLGDYGGTG-YPAGKGGVNGLTLAIAAELKE
MLBr_00862|M.leprae_Br4923 QRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAELDA
. * ::*..* . . . * : *.. .: : **
Mb2788c|M.bovis_AF2122/97 R-IRVNAICPGVVRTRLAEALWKDHEDPLAATIALGRIG--------EPA
Rv2766c|M.tuberculosis_H37Rv R-IRVNAICPGVVRTRLAEALWKDHEDPLAATIALGRIG--------EPA
MMAR_2633|M.marinum_M RGVRANVICPGARTRLSSGPDYEAHINELHRRGLLDEMTRQASLDAPPAD
MUL_3108|M.ulcerans_Agy99 RGVRANVICPGARTRLSSGPNYEAHINELHRRGLLDEMTRQASLDAPPAD
MAB_1210c|M.abscessus_ATCC_199 SGVRANVICPGARTRLSEGPEYTAHLHDLHTRGLLDDMTLQASLVPAPPA
MAV_2962|M.avium_104 YGVRANVVCPGARTRLSTGADYERHIEDLHRRGLLDDMTRRASLDSAPPV
Mflv_1591|M.gilvum_PYR-GCK HGVRANVVCPGAKTRLSTGPEYESHITELNRRGLLDDVSFQGALDAAPPE
Mvan_5166|M.vanbaalenii_PYR-1 HGIRANVVCPGAKTRLSTGSDYESHIAELNRRGMLDDVSYQGALDAAPPE
MSMEG_5869|M.smegmatis_MC2_155 HKVRANVVCPGAKTRLSTGPEYERHIAELNRRGLLDDVSAQGALDAAAPE
TH_1252|M.thermoresistible__bu HSVRANVVCPGARTRLSTGPEYEAHIAELHRRGLLDEVSMQGALDAAPPE
MLBr_00862|M.leprae_Br4923 VGVGLTTICPGVIDTNIIQSTRIDTPTAKQER-VRDRRGQLDMMFRLRRY
: ..:***. . .
Mb2788c|M.bovis_AF2122/97 DIASAVAFLVSDAASWITGETMIIDGGLL------LGNALGFRAAPSTEH
Rv2766c|M.tuberculosis_H37Rv DIASAVAFLVSDAASWITGETMIIDGGLL------LGNALGFRAAPSTEH
MMAR_2633|M.marinum_M FVAPLYAYLASDLASGITGQIFVASGGFVGTFDRPTPRVLGWRDHRADPP
MUL_3108|M.ulcerans_Agy99 FVAPLYAYLASDLASGITGQIFVASGGSVGTFDRPTPRVLGWRDHRADPP
MAB_1210c|M.abscessus_ATCC_199 CVAPLYAYLAGDLSARVTGQIFLAAGGFIGRFKRQTPVPLGYRDHALDAP
MAV_2962|M.avium_104 FVAPVYGYLVSDLARDVTGQILVAAGGFVGSFDRQTPRLLGYRDHHRAGP
Mflv_1591|M.gilvum_PYR-GCK YVAPTYAYLASDLATQVTGQIFIAAGGFVGRFERQTPSLVAYRGHDDEPP
Mvan_5166|M.vanbaalenii_PYR-1 YAAPTYAYLVSDLANHITGQIFIAAGGFVGRFDRQTPALVAYRGHHDEPP
MSMEG_5869|M.smegmatis_MC2_155 YAAATYAYLVSDAARDVTGRIFIAAGGFVGEFARPTPGFLGFRDHHTSPP
TH_1252|M.thermoresistible__bu YVAPTYAYLVSDLAKDITGRIFIAAGGFLGEFARPTPDFLGFRDANDSPP
MLBr_00862|M.leprae_Br4923 GPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTARIRVI-
: . . . . . : :
Mb2788c|M.bovis_AF2122/97 ------------------
Rv2766c|M.tuberculosis_H37Rv ------------------
MMAR_2633|M.marinum_M WPVEDLHTMINARLLDTD
MUL_3108|M.ulcerans_Agy99 WPVEDLHTMINARLLDTD
MAB_1210c|M.abscessus_ATCC_199 WSMEEIDDLMGRR-----
MAV_2962|M.avium_104 WSIEEVHTMIGAAATP--
Mflv_1591|M.gilvum_PYR-GCK WTVEEIAAQVR-------
Mvan_5166|M.vanbaalenii_PYR-1 WTVEEIAAQVG-------
MSMEG_5869|M.smegmatis_MC2_155 WTADELHQMISGG-----
TH_1252|M.thermoresistible__bu FSVEEIHDMINR------
MLBr_00862|M.leprae_Br4923 ------------------