For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVVGGTRGVGLAVAELLAALGAGVVVNGRDEVAAGQAAERIPRAIGFSGSPSDPDVADALIDTCVREFGR IDILVNCAGTAEPVGSSILNVTPAQFQELIDAHLGTAFQTCRAAAPLMARQGSGAIVNTSSFAFLGDYGG TGYAAGKGAVNSLTLAIAAELKEHGIRANVVCPGAKTRLSTGSDYESHIAELNRRGMLDDVSYQGALDAA PPEYAAPTYAYLVSDLANHITGQIFIAAGGFVGRFDRQTPALVAYRGHHDEPPWTVEEIAAQVG
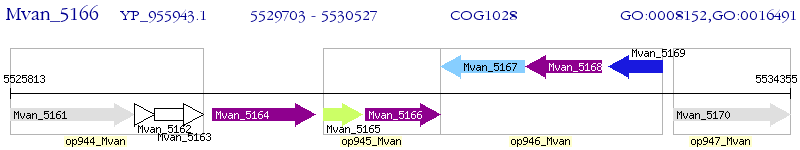
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5166 | - | - | 100% (274) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4960 | - | 9e-26 | 31.34% (284) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_1975 | fabG | 6e-24 | 32.80% (250) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2788c | fabG | 1e-27 | 33.60% (247) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1591 | - | 1e-120 | 83.53% (249) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2766c | fabG | 1e-27 | 33.60% (247) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 7e-16 | 28.34% (247) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_1210c | - | 9e-87 | 57.19% (278) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_2633 | - | 3e-98 | 61.01% (277) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_2962 | - | 1e-101 | 63.67% (278) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_5869 | - | 1e-114 | 74.72% (269) | putative short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1252 | - | 1e-103 | 73.58% (246) | putative short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_3108 | - | 2e-95 | 59.57% (277) | short-chain type dehydrogenase/reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5166|M.vanbaalenii_PYR-1 ------------------------MVVGGTRGVGLAVAELLAALGAGVVV
Mflv_1591|M.gilvum_PYR-GCK ------------------------------------------------MV
MSMEG_5869|M.smegmatis_MC2_155 ------------------------MVVGGSRGIGLAVAGLLAAQGAGVVV
TH_1252|M.thermoresistible__bu -----------------------------------------------VVV
MMAR_2633|M.marinum_M --------MTDRD----LLAGRGVVVVGGSRGIGRAVAELLARLGAGVVI
MUL_3108|M.ulcerans_Agy99 --------MTDRD----LLAGRGVVVVGGSRGIGRAVAELLARLGAGVVI
MAV_2962|M.avium_104 --------MAAVSGAAALLSGRGAVVSGGSRGIGRAVAELLAGLGAGVVV
MAB_1210c|M.abscessus_ATCC_199 --------------------MPNCVVVGGSRGIGRAVALLLGELGTNVVV
Mb2788c|M.bovis_AF2122/97 -------------MTSLDLTGRTAIITGASRGIGLAIAQQLAAAGAHVVL
Rv2766c|M.tuberculosis_H37Rv -------------MTSLDLTGRTAIITGASRGIGLAIAQQLAAAGAHVVL
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
:
Mvan_5166|M.vanbaalenii_PYR-1 NGRDEVAAGQAAERIPR----AIGFSGSPSDPDVADALIDTCVREFGRID
Mflv_1591|M.gilvum_PYR-GCK NGRDPDAAHEAAQRIPG----ALAHPGSPADPAVADALIDICVREFGRID
MSMEG_5869|M.smegmatis_MC2_155 NGRDADAAEAAAKRIS-----GVAHAGSPADPAVADALVDTCVTEFGRID
TH_1252|M.thermoresistible__bu NGRDPEVVTEAAQRIS-----GVGHAGSPADPAVADALVDTCVQNFGSID
MMAR_2633|M.marinum_M SGREPDAVRDATEAVISAGGRAAGIAGAADHEDTANALVDRCMTSFGRLD
MUL_3108|M.ulcerans_Agy99 SGREPDAVRDATEAVISAGGRAAGIAGAADHEDTANALVDRCMTSFGRLD
MAV_2962|M.avium_104 NGRDPQAVRETVAAITAAGGRATAVVGAADDERIARSLVDECIGAFGRLD
MAB_1210c|M.abscessus_ATCC_199 NGRHADVVDDTVGAITSSGGTAVGFVGAPDNERTATDLIDTCRTQYGGID
Mb2788c|M.bovis_AF2122/97 TARRQEAADEAAAQVGDR---ALGVGAHAVDEDAARRCVDLTLERFGSVD
Rv2766c|M.tuberculosis_H37Rv TARRQEAADEAAAQVGDR---ALGVGAHAVDEDAARRCVDLTLERFGSVD
MLBr_01807|M.leprae_Br4923 THR--------ASVVPDW---FFGVECDVTDNDAIGAAFKAVEEHQGPVE
. * . : . . .. * ::
Mvan_5166|M.vanbaalenii_PYR-1 ILVNCAGTAEPVGSSILNVTPAQFQELIDAHLGTAFQTCRAAAPLMARQG
Mflv_1591|M.gilvum_PYR-GCK ILINCAGTAEPRGSSILNVTSAQFRELLDAHLGTTFETCRAAAPVMVAQG
MSMEG_5869|M.smegmatis_MC2_155 ILVNCAGTAEPHGSSILDITSAQFRELLDAHLGTVFETCRAAAPKMVAHG
TH_1252|M.thermoresistible__bu ILINCAGTAEPPGSSILNISSGQFRELLDAHLGTVFETCRAAAPRMVAQG
MMAR_2633|M.marinum_M ALINCAGIAEPPGSSILNISPAQFDGLLRAHLGTVFHTCRVAAPVLAAQG
MUL_3108|M.ulcerans_Agy99 ALINCAGIAEPPGSSILNISPAQSDGLLRAHLGTVFHACRVAAPVLAAQG
MAV_2962|M.avium_104 ALINCAGIAEPAGSSILNITADEFDHLIGAHLGTAFHTCRAAAPVMVEQR
MAB_1210c|M.abscessus_ATCC_199 ALINCAGIPEPTGSSILTITSGDFHRLIDAHLGTVFHTCRVVAPLMVQQR
Mb2788c|M.bovis_AF2122/97 ILINNAGTN-PAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMGEH
Rv2766c|M.tuberculosis_H37Rv ILINNAGTN-PAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMGEH
MLBr_01807|M.leprae_Br4923 VLVANAGIT-SDM-LIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR
*: ** . :: :: .:* : . .
Mvan_5166|M.vanbaalenii_PYR-1 SGAIVNTSSFAFLGDYGGTG-YAAGKGAVNSLTLAIAAELKEHGIRANVV
Mflv_1591|M.gilvum_PYR-GCK GGAIVNTGSFAFLGDYGGTG-YPAGKGAVNSLTLAIAAELAEHGVRANVV
MSMEG_5869|M.smegmatis_MC2_155 GGAIVNTGSFAYLGDYGGTG-YPAGKGGVNALTMAIAAELREHKVRANVV
TH_1252|M.thermoresistible__bu GGAIVNTSSFAFLGDYGGTG-YPAGKGGVNGLTLAIAAELKEHSVRANVV
MMAR_2633|M.marinum_M HGSIINTGSVAFLGDYGGSG-YPAGKAAVNGLTLAISAELKARGVRANVI
MUL_3108|M.ulcerans_Agy99 HGSIINTGSVAFLGDYGGSG-YPAGKAAVNGLTLAISAELKARGVRANVI
MAV_2962|M.avium_104 HGSIVNTSSVAFLGDYGGTG-YPAGKGAVNALTMAIAAELKAYGVRANVV
MAB_1210c|M.abscessus_ATCC_199 HGAIINTGSTASLGIYGGTG-YPAGKAAVNGLTLAIAAELKRSGVRANVI
Mb2788c|M.bovis_AF2122/97 GGAVVNTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-IRVNAI
Rv2766c|M.tuberculosis_H37Rv GGAVVNTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-IRVNAI
MLBr_01807|M.leprae_Br4923 FGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITANVV
* : .* : . . * * *..: :: :: ** : .*.:
Mvan_5166|M.vanbaalenii_PYR-1 CPGAKTRLSTGSDYESHIAELNRRGMLDDVSYQGALDAAPPEYAAPTYAY
Mflv_1591|M.gilvum_PYR-GCK CPGAKTRLSTGPEYESHITELNRRGLLDDVSFQGALDAAPPEYVAPTYAY
MSMEG_5869|M.smegmatis_MC2_155 CPGAKTRLSTGPEYERHIAELNRRGLLDDVSAQGALDAAAPEYAAATYAY
TH_1252|M.thermoresistible__bu CPGARTRLSTGPEYEAHIAELHRRGLLDEVSMQGALDAAPPEYVAPTYAY
MMAR_2633|M.marinum_M CPGARTRLSSGPDYEAHINELHRRGLLDEMTRQASLDAPPADFVAPLYAY
MUL_3108|M.ulcerans_Agy99 CPGARTRLSSGPNYEAHINELHRRGLLDEMTRQASLDAPPADFVAPLYAY
MAV_2962|M.avium_104 CPGARTRLSTGADYERHIEDLHRRGLLDDMTRRASLDSAPPVFVAPVYGY
MAB_1210c|M.abscessus_ATCC_199 CPGARTRLSEGPEYTAHLHDLHTRGLLDDMTLQASLVPAPPACVAPLYAY
Mb2788c|M.bovis_AF2122/97 CPGVVRTRLAEALWKDHEDPLAATIALGRIG--------EPADIASAVAF
Rv2766c|M.tuberculosis_H37Rv CPGVVRTRLAEALWKDHEDPLAATIALGRIG--------EPADIASAVAF
MLBr_01807|M.leprae_Br4923 APGFIDTEMTRAMDSKYQEAALQAIPLGRIG--------AVEEIAGAVSF
.** . : *. : * .:
Mvan_5166|M.vanbaalenii_PYR-1 LVSDLANHITGQIFIAAGGFVGRFDRQTPALVAYRGHHDEPPWTVEEIAA
Mflv_1591|M.gilvum_PYR-GCK LASDLATQVTGQIFIAAGGFVGRFERQTPSLVAYRGHDDEPPWTVEEIAA
MSMEG_5869|M.smegmatis_MC2_155 LVSDAARDVTGRIFIAAGGFVGEFARPTPGFLGFRDHHTSPPWTADELHQ
TH_1252|M.thermoresistible__bu LVSDLAKDITGRIFIAAGGFLGEFARPTPDFLGFRDANDSPPFSVEEIHD
MMAR_2633|M.marinum_M LASDLASGITGQIFVASGGFVGTFDRPTPRVLGWRDHRADPPWPVEDLHT
MUL_3108|M.ulcerans_Agy99 LASDLASGITGQIFVASGGSVGTFDRPTPRVLGWRDHRADPPWPVEDLHT
MAV_2962|M.avium_104 LVSDLARDVTGQILVAAGGFVGSFDRQTPRLLGYRDHHRAGPWSIEEVHT
MAB_1210c|M.abscessus_ATCC_199 LAGDLSARVTGQIFLAAGGFIGRFKRQTPVPLGYRDHALDAPWSMEEIDD
Mb2788c|M.bovis_AF2122/97 LVSDAASWITGETMIIDGG----------LLLGNALGFRAAPSTEH----
Rv2766c|M.tuberculosis_H37Rv LVSDAASWITGETMIIDGG----------LLLGNALGFRAAPSTEH----
MLBr_01807|M.leprae_Br4923 LASEDAGYISGAVIPVDGG----------LGMGH----------------
*..: : ::* : ** :.
Mvan_5166|M.vanbaalenii_PYR-1 QVG-------
Mflv_1591|M.gilvum_PYR-GCK QVR-------
MSMEG_5869|M.smegmatis_MC2_155 MISGG-----
TH_1252|M.thermoresistible__bu MINR------
MMAR_2633|M.marinum_M MINARLLDTD
MUL_3108|M.ulcerans_Agy99 MINARLLDTD
MAV_2962|M.avium_104 MIGAAATP--
MAB_1210c|M.abscessus_ATCC_199 LMGRR-----
Mb2788c|M.bovis_AF2122/97 ----------
Rv2766c|M.tuberculosis_H37Rv ----------
MLBr_01807|M.leprae_Br4923 ----------