For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVVGGSRGIGLAVAGLLAAQGAGVVVNGRDADAAEAAAKRISGVAHAGSPADPAVADALVDTCVTEFGRI DILVNCAGTAEPHGSSILDITSAQFRELLDAHLGTVFETCRAAAPKMVAHGGGAIVNTGSFAYLGDYGGT GYPAGKGGVNALTMAIAAELREHKVRANVVCPGAKTRLSTGPEYERHIAELNRRGLLDDVSAQGALDAAA PEYAAATYAYLVSDAARDVTGRIFIAAGGFVGEFARPTPGFLGFRDHHTSPPWTADELHQMISGG
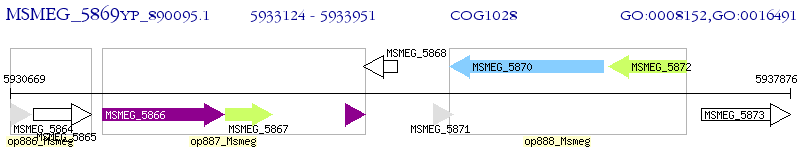
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5869 | - | - | 100% (275) | putative short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2598 | - | 9e-29 | 32.93% (249) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4358 | - | 5e-24 | 33.20% (241) | D-beta-hydroxybutyrate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 2e-28 | 32.53% (249) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1591 | - | 1e-111 | 79.59% (245) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2857c | - | 2e-28 | 32.53% (249) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 9e-16 | 32.70% (211) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1210c | - | 9e-84 | 56.68% (277) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_2633 | - | 3e-94 | 60.71% (280) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_2962 | - | 8e-96 | 62.82% (277) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_1252 | - | 1e-118 | 81.20% (250) | putative short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_3108 | - | 1e-91 | 59.64% (280) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_5166 | - | 1e-114 | 74.72% (269) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5869|M.smegmatis_MC2_155 --------------------------------------------------
TH_1252|M.thermoresistible__bu --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2633|M.marinum_M --------------------------------------------------
MUL_3108|M.ulcerans_Agy99 --------------------------------------------------
MAV_2962|M.avium_104 --------------------------------------------------
MAB_1210c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mb2882c|M.bovis_AF2122/97 ------------------MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRA
Rv2857c|M.tuberculosis_H37Rv ------------------MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRA
MSMEG_5869|M.smegmatis_MC2_155 -------------------------------MVVGGSRGIGLAVAGLLAA
TH_1252|M.thermoresistible__bu --------------------------------------------------
Mflv_1591|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5166|M.vanbaalenii_PYR-1 -------------------------------MVVGGTRGVGLAVAELLAA
MMAR_2633|M.marinum_M ---------------MTDRD----LLAGRGVVVVGGSRGIGRAVAELLAR
MUL_3108|M.ulcerans_Agy99 ---------------MTDRD----LLAGRGVVVVGGSRGIGRAVAELLAR
MAV_2962|M.avium_104 ---------------MAAVSGAAALLSGRGAVVSGGSRGIGRAVAELLAG
MAB_1210c|M.abscessus_ATCC_199 ---------------------------MPNCVVVGGSRGIGRAVALLLGE
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
Mb2882c|M.bovis_AF2122/97 EGATIVVGDVDVEAGGAAADELSG-----LFVPTDVCDEDAVNGLFDGAA
Rv2857c|M.tuberculosis_H37Rv EGATIVVGDVDVEAGGAAADELSG-----LFVPTDVCDEDAVNGLFDGAA
MSMEG_5869|M.smegmatis_MC2_155 QGAGVVVNGRDADAAEAAAKRIS-----GVAHAGSPADPAVADALVDTCV
TH_1252|M.thermoresistible__bu ----VVVNGRDPEVVTEAAQRIS-----GVGHAGSPADPAVADALVDTCV
Mflv_1591|M.gilvum_PYR-GCK -----MVNGRDPDAAHEAAQRIPG----ALAHPGSPADPAVADALIDICV
Mvan_5166|M.vanbaalenii_PYR-1 LGAGVVVNGRDEVAAGQAAERIPR----AIGFSGSPSDPDVADALIDTCV
MMAR_2633|M.marinum_M LGAGVVISGREPDAVRDATEAVISAGGRAAGIAGAADHEDTANALVDRCM
MUL_3108|M.ulcerans_Agy99 LGAGVVISGREPDAVRDATEAVISAGGRAAGIAGAADHEDTANALVDRCM
MAV_2962|M.avium_104 LGAGVVVNGRDPQAVRETVAAITAAGGRATAVVGAADDERIARSLVDECI
MAB_1210c|M.abscessus_ATCC_199 LGTNVVVNGRHADVVDDTVGAITSSGGTAVGFVGAPDNERTATDLIDTCR
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
::.. . . :. : . . : :
Mb2882c|M.bovis_AF2122/97 ETYGRIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAAL
Rv2857c|M.tuberculosis_H37Rv ETYGRIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAAL
MSMEG_5869|M.smegmatis_MC2_155 TEFGRIDILVNCAGTAEPHGSSILDITSAQFRELLDAHLGTVFETCRAAA
TH_1252|M.thermoresistible__bu QNFGSIDILINCAGTAEPPGSSILNISSGQFRELLDAHLGTVFETCRAAA
Mflv_1591|M.gilvum_PYR-GCK REFGRIDILINCAGTAEPRGSSILNVTSAQFRELLDAHLGTTFETCRAAA
Mvan_5166|M.vanbaalenii_PYR-1 REFGRIDILVNCAGTAEPVGSSILNVTPAQFQELIDAHLGTAFQTCRAAA
MMAR_2633|M.marinum_M TSFGRLDALINCAGIAEPPGSSILNISPAQFDGLLRAHLGTVFHTCRVAA
MUL_3108|M.ulcerans_Agy99 TSFGRLDALINCAGIAEPPGSSILNISPAQSDGLLRAHLGTVFHACRVAA
MAV_2962|M.avium_104 GAFGRLDALINCAGIAEPAGSSILNITADEFDHLIGAHLGTAFHTCRAAA
MAB_1210c|M.abscessus_ATCC_199 TQYGGIDALINCAGIPEPTGSSILTITSGDFHRLIDAHLGTVFHTCRVVA
MLBr_00862|M.leprae_Br4923 AKHGVPDIVVNNAGIG--QAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFG
.* * .* ** . : : .:* . **.
Mb2882c|M.bovis_AF2122/97 RHMVLAGKG-SIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFA
Rv2857c|M.tuberculosis_H37Rv RHMVLAGKG-SIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFA
MSMEG_5869|M.smegmatis_MC2_155 PKMVAHGGG-AIVNTGSFAYLG--DYGGTGYPAGKGGVNALTMAIAAELR
TH_1252|M.thermoresistible__bu PRMVAQGGG-AIVNTSSFAFLG--DYGGTGYPAGKGGVNGLTLAIAAELK
Mflv_1591|M.gilvum_PYR-GCK PVMVAQGGG-AIVNTGSFAFLG--DYGGTGYPAGKGAVNSLTLAIAAELA
Mvan_5166|M.vanbaalenii_PYR-1 PLMARQGSG-AIVNTSSFAFLG--DYGGTGYAAGKGAVNSLTLAIAAELK
MMAR_2633|M.marinum_M PVLAAQGHG-SIINTGSVAFLG--DYGGSGYPAGKAAVNGLTLAISAELK
MUL_3108|M.ulcerans_Agy99 PVLAAQGHG-SIINTGSVAFLG--DYGGSGYPAGKAAVNGLTLAISAELK
MAV_2962|M.avium_104 PVMVEQRHG-SIVNTSSVAFLG--DYGGTGYPAGKGAVNALTMAIAAELK
MAB_1210c|M.abscessus_ATCC_199 PLMVQQRHG-AIINTGSTASLG--IYGGTGYPAGKAAVNGLTLAIAAELK
MLBr_00862|M.leprae_Br4923 QRLVERGTGGHIVNVSSMAAYAP-LQSLSAYCTSKAATYMFSDCLRAELD
:. * *:*..* . . .* :.*... :: : .::
Mb2882c|M.bovis_AF2122/97 RQGIRVNALCPGPVNT--PLLQELFAKNPERAARR-----MVHVPLGRFA
Rv2857c|M.tuberculosis_H37Rv RQGIRVNALCPGPVNT--PLLQELFAKNPERAARR-----MVHVPLGRFA
MSMEG_5869|M.smegmatis_MC2_155 EHKVRANVVCPGAKTR--LSTGPEYERHIAELNRRGLLDDVSAQGALDAA
TH_1252|M.thermoresistible__bu EHSVRANVVCPGARTR--LSTGPEYEAHIAELHRRGLLDEVSMQGALDAA
Mflv_1591|M.gilvum_PYR-GCK EHGVRANVVCPGAKTR--LSTGPEYESHITELNRRGLLDDVSFQGALDAA
Mvan_5166|M.vanbaalenii_PYR-1 EHGIRANVVCPGAKTR--LSTGSDYESHIAELNRRGMLDDVSYQGALDAA
MMAR_2633|M.marinum_M ARGVRANVICPGARTR--LSSGPDYEAHINELHRRGLLDEMTRQASLDAP
MUL_3108|M.ulcerans_Agy99 ARGVRANVICPGARTR--LSSGPNYEAHINELHRRGLLDEMTRQASLDAP
MAV_2962|M.avium_104 AYGVRANVVCPGARTR--LSTGADYERHIEDLHRRGLLDDMTRRASLDSA
MAB_1210c|M.abscessus_ATCC_199 RSGVRANVICPGARTR--LSEGPEYTAHLHDLHTRGLLDDMTLQASLVPA
MLBr_00862|M.leprae_Br4923 AVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRLRRY
: ..:*** * :
Mb2882c|M.bovis_AF2122/97 EPDEIAAAVAFLASDDASFITASTFLVDGGISSAYVTPL-----------
Rv2857c|M.tuberculosis_H37Rv EPDEIAAAVAFLASDDASFITASTFLVDGGISSAYVTPL-----------
MSMEG_5869|M.smegmatis_MC2_155 APEYAAATYAYLVSDAARDVTGRIFIAAGGFVGEFARPTPGFLGFRDHHT
TH_1252|M.thermoresistible__bu PPEYVAPTYAYLVSDLAKDITGRIFIAAGGFLGEFARPTPDFLGFRDAND
Mflv_1591|M.gilvum_PYR-GCK PPEYVAPTYAYLASDLATQVTGQIFIAAGGFVGRFERQTPSLVAYRGHDD
Mvan_5166|M.vanbaalenii_PYR-1 PPEYAAPTYAYLVSDLANHITGQIFIAAGGFVGRFDRQTPALVAYRGHHD
MMAR_2633|M.marinum_M PADFVAPLYAYLASDLASGITGQIFVASGGFVGTFDRPTPRVLGWRDHRA
MUL_3108|M.ulcerans_Agy99 PADFVAPLYAYLASDLASGITGQIFVASGGSVGTFDRPTPRVLGWRDHRA
MAV_2962|M.avium_104 PPVFVAPVYGYLVSDLARDVTGQILVAAGGFVGSFDRQTPRLLGYRDHHR
MAB_1210c|M.abscessus_ATCC_199 PPACVAPLYAYLAGDLSARVTGQIFLAAGGFIGRFKRQTPVPLGYRDHAL
MLBr_00862|M.leprae_Br4923 GPDKVADT---ILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTARIR
. * : . . . .
Mb2882c|M.bovis_AF2122/97 ---------------------
Rv2857c|M.tuberculosis_H37Rv ---------------------
MSMEG_5869|M.smegmatis_MC2_155 SPPWTADELHQMISGG-----
TH_1252|M.thermoresistible__bu SPPFSVEEIHDMINR------
Mflv_1591|M.gilvum_PYR-GCK EPPWTVEEIAAQVR-------
Mvan_5166|M.vanbaalenii_PYR-1 EPPWTVEEIAAQVG-------
MMAR_2633|M.marinum_M DPPWPVEDLHTMINARLLDTD
MUL_3108|M.ulcerans_Agy99 DPPWPVEDLHTMINARLLDTD
MAV_2962|M.avium_104 AGPWSIEEVHTMIGAAATP--
MAB_1210c|M.abscessus_ATCC_199 DAPWSMEEIDDLMGRR-----
MLBr_00862|M.leprae_Br4923 VI-------------------