For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GGPVTRFDSAAIAEWRRYGTVPLAAGLGYSMGVIHVYSLGAFMGPLQDEFGWSRAQASVGLTIVGMAAAV AALPIGLMVDRLGPRRVGIIGAGLMAGAFALLGTATGTMGNWILLWSVLAFACFWVQTTVWTSAVASRFE TSRGLAFAVTLSGGSLAAAVFPPLATLLIGEWGWRGALFGLGGLWGGLVLIVVLLFFRGAQDDKKPAPAR TVIAPTAAAADLPGVTLAEGLRSATFYKLLVAAGLFAFTTLGIVVHLVPILRDAGAEPLAAAGTAALVGI FSIVGRLGVGLLLDRFPGRVVGACAYLVPILGCAALLIDGSSPLSQTVAAALFGLTLGAEVDVIAYLASR YFGLRNFGGLFGGLVAALSLGTAFGPLAAGATSDHFGGYTNFLILTMVLMGLSSMALASLKPPPVWTVTS PGEDARADRFPQP*
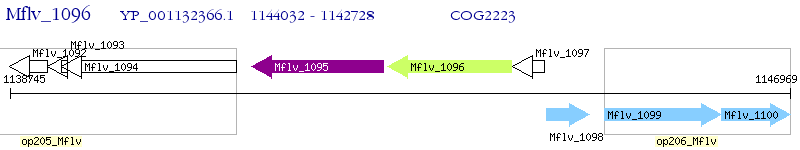
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1096 | - | - | 100% (434) | major facilitator transporter |
| M. gilvum PYR-GCK | Mflv_2728 | - | 3e-09 | 24.19% (401) | major facilitator transporter |
| M. gilvum PYR-GCK | Mflv_2530 | - | 4e-09 | 21.69% (415) | major facilitator transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1288c | - | 2e-09 | 22.93% (375) | integral membrane transport protein |
| M. tuberculosis H37Rv | Rv1258c | - | 2e-09 | 22.93% (375) | integral membrane transport protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4839c | - | 8e-22 | 25.51% (396) | MFS family transporter |
| M. marinum M | MMAR_3642 | - | 2e-06 | 27.17% (184) | integral membrane transport protein |
| M. avium 104 | MAV_4712 | - | 4e-07 | 25.74% (373) | transporter, major facilitator family protein |
| M. smegmatis MC2 155 | MSMEG_4147 | - | 1e-42 | 32.26% (403) | major facilitator superfamily protein MFS_1, putative |
| M. thermoresistible (build 8) | TH_0557 | - | 1e-26 | 28.57% (427) | major facilitator superfamily protein, putative |
| M. ulcerans Agy99 | MUL_1468 | - | 2e-06 | 27.37% (190) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_3219 | - | 1e-20 | 25.76% (396) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4839c|M.abscessus_ATCC_199 ------MSTAAVHAQPHSITGLRGVLITLCLTEITSWGVLYYAFPVLAPR
Mvan_3219|M.vanbaalenii_PYR-1 ------MAESDRADR-------RSVLLTLCLTEITSWGVLYYAFSVLSPA
Mb1288c|M.bovis_AF2122/97 ------MRNSN-----------RGPAFLILFATLMAAAGDGVSI-VAFPW
Rv1258c|M.tuberculosis_H37Rv ------MRNSN-----------RGPAFLILFATLMAAAGDGVSI-VAFPW
MMAR_3642|M.marinum_M ------MNRTQ-----------LLTIFATGLGLFMIFLDAFIVN-VALPD
MUL_1468|M.ulcerans_Agy99 ------MNRTQ-----------LLTIFATGLGLFMIFLDAFIVN-VALPD
Mflv_1096|M.gilvum_PYR-GCK ----MGGPVTRFDSAAIAEWRRYGTVPLAAGLGYSMGVIHVYSLGAFMGP
MSMEG_4147|M.smegmatis_MC2_155 MSAASHDVMTPPGAIGEFRRSWAALTAVTVGIAIGVGVLPTYVNGLVIPE
TH_0557|M.thermoresistible__bu -------VTAAPSYLGEVRVNWRPLAAATAGLSAGLS-LSAYTNAAMAPQ
MAV_4712|M.avium_104 ------MTDSSPPAKGTFTVEMLSRAKRGVTAVFVAHGLLFASWAAHIPQ
:
MAB_4839c|M.abscessus_ATCC_199 ITADTGWSAPAVTAAFSAALVVAAAAGIGVGRWLDRHGPRWLMTAGSLWG
Mvan_3219|M.vanbaalenii_PYR-1 ICADTGWSAPAVTAAFSAGLVISALLGIPVGRWLDRHGPRWIMTGGSVLG
Mb1288c|M.bovis_AF2122/97 LVLQREGSAGQASIVASATMLPLLFATLVAGTAVDYFGRRRVSMVADALS
Rv1258c|M.tuberculosis_H37Rv LVLQREGSAGQASIVASATMLPLLFATLVAGTAVDYFGRRRVSMVADALS
MMAR_3642|M.marinum_M IQNTFGVGEDGLQWVVAAYSLGMAVFIMSAATLADRYGRRRWYLMGIALF
MUL_1468|M.ulcerans_Agy99 IQNTFGVGEDGLQWVVAAYSLGMAVFIMSAATLADRYGRRRWYLMGIVLF
Mflv_1096|M.gilvum_PYR-GCK LQDEFGWSRAQASVGLTIVGMAAAVAALPIGLMVDRLGPRRVGIIGAGLM
MSMEG_4147|M.smegmatis_MC2_155 LEAEFGWSRTALSALPLIGSLIIIATAPVVGVVVDRFGVRIPAVSSLVAF
TH_0557|M.thermoresistible__bu FLAAFDWSRSDFALTG-VIALLTFVFLPVYGRLTDLFGVRRIALIGVTGL
MAV_4712|M.avium_104 VKAGLGLDDAALGTALFGAPLGSVLATLAGHWALPRWGSHRLIPVTVAGY
. . : * :
MAB_4839c|M.abscessus_ATCC_199 PAALIGVAMAPN---------YGWFLAAWLMAGIAMAAVFYPPAFAALTR
Mvan_3219|M.vanbaalenii_PYR-1 SIALAGVAAAPD---------VVSFTAAWVVAGVAMSAVFYAPAFAALTR
Mb1288c|M.bovis_AF2122/97 GAAVAGVPLVAWGYGGDAVNVLVLAVLAALAAAFGPAGMTARDSMLPEAA
Rv1258c|M.tuberculosis_H37Rv GAAVAGVPLVAWGYGGDAVNVLVLAVLAALAAAFGPAGMTARDSMLPEAA
MMAR_3642|M.marinum_M TLGSIACGLAPS---------IAMLTAARGIQGFGAATVSVTSLALVSAA
MUL_1468|M.ulcerans_Agy99 TLGSIACGLAPS---------IAMLTAARGVQGFGAATVSVTSLALVSAA
Mflv_1096|M.gilvum_PYR-GCK AGAFALLGTATG--------TMGNWILLWSVLAFACFWVQTTVWTSAVAS
MSMEG_4147|M.smegmatis_MC2_155 AAGYFLLAYSNS--------SFGQYLLMFIVMYLLAAASTAVAFTRTINE
TH_0557|M.thermoresistible__bu PVCWIGYALMSG--------PIWQYFAIHVAMIALGTTTTPAIYSRLVAI
MAV_4712|M.avium_104 AAAGTTVGLARS-----------GPALFAALALWGMFQGALDVAMNTQAG
MAB_4839c|M.abscessus_ATCC_199 WYGP-GAVRALMVLTLVAGLASTVFVPLTAVLASHMDWRTTYLALAVLLT
Mvan_3219|M.vanbaalenii_PYR-1 YFRP-DAVRALTALTLAAGLASTVFAPLTAAISAHMDWRETYLVLAAAMA
Mb1288c|M.bovis_AF2122/97 ARAGWSLDRINGAYEAILNLAFIVGPAIGGLMIATVGGITTMWITATAFG
Rv1258c|M.tuberculosis_H37Rv ARAGWSLDRINGAYEAILNLAFIVGPAIGGLMIATVGGITTMWITATAFG
MMAR_3642|M.marinum_M FPNPKEKARAIGIWTAVASVGTATGPTLGGLLVDTWGWRSIFYVNVPMGV
MUL_1468|M.ulcerans_Agy99 FPNPKEKARAIGIWTAVASVGTATGPTLGGLLVDTWGWRSIFYVNVPMGV
Mflv_1096|M.gilvum_PYR-GCK RFET-SRGLAFAVTLSGGSLAAAVFPPLATLLIGEWGWRGALFGLGGLWG
MSMEG_4147|M.smegmatis_MC2_155 RYDR-ARGLALGIALSGAGVVGFLVPSVVGPVIADN-WRTGYRILAVVIL
TH_0557|M.thermoresistible__bu RFDR-ARGLALAVAISGPPLLGALGVPALDAVNQTLGWRWGCVAIAVAIA
MAV_4712|M.avium_104 TVERRAGAPMMARFHGMWSLGTLAGALIGAACVGAGIGLTAQLTVLGAVV
:
MAB_4839c|M.abscessus_ATCC_199 IITVPAHIIGLRR-------------PWPSLPATHLAESPT---RIARSR
Mvan_3219|M.vanbaalenii_PYR-1 VITVPAHFFGLRR-------------PWPHIEHHHLVEPPT---RIARSR
Mb1288c|M.bovis_AF2122/97 LSILAIAALQLEG----------AGKPHHTSRPQGLVSGIAEGLRFVWNL
Rv1258c|M.tuberculosis_H37Rv LSILAIAALQLEG----------AGKPHHTSRPQGLVSGIAEGLRFVWNL
MMAR_3642|M.marinum_M FVILLTVGFVAESRNARAGRLDLSGQALFIVTVGAFVYAVIEGPQIGWTS
MUL_1468|M.ulcerans_Agy99 FVILLTVGFVAESRNARAGQLDLSGQALFIVTVGAFVYAVIEGPQIGWTS
Mflv_1096|M.gilvum_PYR-GCK GLVLIVVLLFFRG-------AQDDKKPAPARTVIAPTAAAADLPGVTLAE
MSMEG_4147|M.smegmatis_MC2_155 LCAVVVAALMPRR------------TAKTAAALPRHPGGRGLVLELIRQP
TH_0557|M.thermoresistible__bu VVGFAALALAPRPNPAADPAPADPTGETPSRGRDDTHRRTGAAYRMIGRN
MAV_4712|M.avium_104 LLVVVMLTRRLLP---------------DAADSVAAPPEPAAGRRVTPAV
. .
MAB_4839c|M.abscessus_ATCC_199 PFISLIIAFALASCASYAVIVN------LVPLMNQRGLDGTVAATALALG
Mvan_3219|M.vanbaalenii_PYR-1 QFVALTVAFAMAGLASYAVIVN------LVPLMTAHGISTQAAALALGLG
Mb1288c|M.bovis_AF2122/97 RVLRTLGMIDLTVTALYLPMES-----VLFPKYFTDHQQPVQLGWALMAI
Rv1258c|M.tuberculosis_H37Rv RVLRTLGMIDLTVTALYLPMES-----VLFPKYFTDHQQPVQLGWALMAI
MMAR_3642|M.marinum_M PLILTLLMTAAVGCALFLWLEGRSADPMMDLTLFRDSSYALSIGTICTVF
MUL_1468|M.ulcerans_Agy99 PLILTLLMTAIVGCALFLWLEGRSADPMMDLTLFRDSSYALSIGTICTVF
Mflv_1096|M.gilvum_PYR-GCK GLRSATFYKLLVAAGLFAFTTLG-IVVHLVPILRDAGAEPLAAAGTAALV
MSMEG_4147|M.smegmatis_MC2_155 RFWRLAIAFLTLALAVGGMLQH------LFPMLRDAGVSAAAAARTASLV
TH_0557|M.thermoresistible__bu PVFWILLAGILLCNLYHSVTTAQ-----LGVVLGDSVGSGNGVAVLVSIF
MAV_4712|M.avium_104 AILAAVSFASFLCEGAATDWSAT-------YLRDVVGAGPSVAAASYAAY
. .
MAB_4839c|M.abscessus_ATCC_199 GVGQVIGRFGYPALAQCISVVPRTLLIMGVVAATTALLGVFASAVS----
Mvan_3219|M.vanbaalenii_PYR-1 GAGQVLGRLGYQRMVRHLSVVPRTALIMAGVAVTTALLGLLTSVAT----
Mb1288c|M.bovis_AF2122/97 AGGGLVGALGYAVLAIRVPRRVTMSTAVLTLGLASMVIAFLPPLP-----
Rv1258c|M.tuberculosis_H37Rv AGGGLVGALGYAVLAIRVPRRVTMSTAVLTLGLASMVIAFLPPLP-----
MMAR_3642|M.marinum_M FA--VYGMLLLTTQFLQNVRGYTPGATGLMILPFSAGVAVVSPLVGSLVA
MUL_1468|M.ulcerans_Agy99 FA--VYGMLLLTTQFLHNVRGYTQGSG------------VVD--------
Mflv_1096|M.gilvum_PYR-GCK GIFSIVGRLGVGLLLDRFPGRVVGACAYLVPILGCAALLIDGSSPLS---
MSMEG_4147|M.smegmatis_MC2_155 GVAVIIARIVVGLLVDRFFAPRVAALVLVVSAAGYVALLAGGPAFA----
TH_0557|M.thermoresistible__bu AAAVIVGRFACGLALDRLPSHLVAAVAMGLPGLGCLLIATPWDGFA----
MAV_4712|M.avium_104 TLTMVVTRFAAGRLHARLPSRRLLPALAVLAVAGMSVTLATADAAAG---
: :
MAB_4839c|M.abscessus_ATCC_199 --------------------------------LIAVAVAAGMARGIMTLL
Mvan_3219|M.vanbaalenii_PYR-1 --------------------------------LITVAVVAGVVRGVMTLL
Mb1288c|M.bovis_AF2122/97 -------------------------------VIMVLCAVVGLVYGPIQPI
Rv1258c|M.tuberculosis_H37Rv -------------------------------VIMVLCAVVGLVYGPIQPI
MMAR_3642|M.marinum_M RVGARVLILIGLGLLMLGLLTLIASEHRSWALVLVGLGFCGTGVALCLTP
MUL_1468|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1096|M.gilvum_PYR-GCK --------------------------------QTVAAALFGLTLGAEVDV
MSMEG_4147|M.smegmatis_MC2_155 ---------------------------------AFAGIGVGLASGAEVDI
TH_0557|M.thermoresistible__bu -------------------------------VLVVAVCCLGAAWGAEGDV
MAV_4712|M.avium_104 ---------------------------------VLGFAALGVG---VALL
MAB_4839c|M.abscessus_ATCC_199 QATAITERWGSTHYGHLSGLLTAPIMLTTALAPFTGAALAQLLHGYPAMF
Mvan_3219|M.vanbaalenii_PYR-1 QATAVTERWGITHYGQLSGILSAPVMLSTALGPFVGAGLATLLGGYGAMF
Mb1288c|M.bovis_AF2122/97 YNYVIQTRAAQHLRGRVVGVMTSLAYAAGPLGLLLAGPLTDAAGLHATFL
Rv1258c|M.tuberculosis_H37Rv YNYVIQTRAAQHLRGRVVGVMTSLAYAAGPLGLLLAGPLTDAAGLHATFL
MMAR_3642|M.marinum_M ITTVAMTAVPPERAGMASGIMSAQRAIGSTIGFAVLG-SVLAAWLSATLE
MUL_1468|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1096|M.gilvum_PYR-GCK IAYLASRYFGLRNFGGLFGGLVAALSLGTAFGPLAAGATSDHFGGYTNFL
MSMEG_4147|M.smegmatis_MC2_155 IGNLTSRYYAIESYGRVFGIFYATFFLGFGSSPLLVAWLRSLTSDYTLPI
TH_0557|M.thermoresistible__bu VAYLVARRFRLTVYSTVLSILSAAIGVSSAVGAVVLSRTLQASDSFNGFL
MAV_4712|M.avium_104 VPTAFSAAYGARGAGSAIAIVAATGWLGYLLGPPLIGHLSEWVGLSGALV
MAB_4839c|M.abscessus_ATCC_199 VALGVAGIIGAAAATATNPSQ-----------------------------
Mvan_3219|M.vanbaalenii_PYR-1 VVLGAVALAAALLATATRVRPVDADG------------------------
Mb1288c|M.bovis_AF2122/97 ALALPIVCTGLVAIRLPALRELDLAPQADIDRPVGSAQ------------
Rv1258c|M.tuberculosis_H37Rv ALALPIVCTGLVAIRLPALRELDLAPQADIDRPVGSAQ------------
MMAR_3642|M.marinum_M PHLEPAVPDPVQRHTIAEVIIDSANPRAHVGGIVPRREIVHRDPVAIAEE
MUL_1468|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1096|M.gilvum_PYR-GCK ILTMVLMGLSSMALASLKPPPVWTVTSPGEDARADRFPQP----------
MSMEG_4147|M.smegmatis_MC2_155 LMSVALMLVAAALLFTAPRFPGEAPADVTNDATVLPAAP-----------
TH_0557|M.thermoresistible__bu VFAGVAAFLGGALFLLSGSRRGESDPPPEGPVRSGTAAVASTG-------
MAV_4712|M.avium_104 TIPVMMTIVAVAIRYTPAFDTADEFHRAPAG-------------------
MAB_4839c|M.abscessus_ATCC_199 ------------------------------------------------
Mvan_3219|M.vanbaalenii_PYR-1 ------------------------------------------------
Mb1288c|M.bovis_AF2122/97 ------------------------------------------------
Rv1258c|M.tuberculosis_H37Rv ------------------------------------------------
MMAR_3642|M.marinum_M DFIEGIRVAFFIATASLGFVFLAGWRWFPRGGAGMLGDAEREADAVGL
MUL_1468|M.ulcerans_Agy99 ------------------------------------------------
Mflv_1096|M.gilvum_PYR-GCK ------------------------------------------------
MSMEG_4147|M.smegmatis_MC2_155 ------------------------------------------------
TH_0557|M.thermoresistible__bu ------------------------------------------------
MAV_4712|M.avium_104 ------------------------------------------------