For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAENRRSGRWRTGQQNKQRILDAARDHFMRSGYEKATVRGIAAEAGVDVAMVYYFFGNKEGLFNAAVLDT PEHPLHQLAALLDDGPEEIGARLVREVIRRWDEGGSFDLLLTVFKSADIHPLARKLLHDSLAGPVAQRVA AEFGVDDAELRVELVTVHMVGLAFARYQLKIEPLASAGLDDLVSWLGPTVQRYLTAPDP
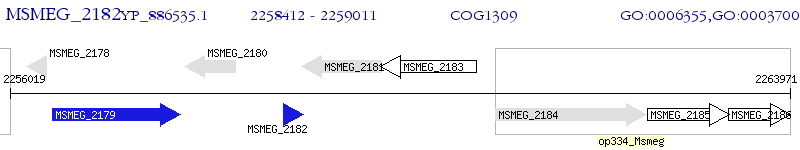
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2182 | - | - | 100% (199) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_3765 | - | 3e-26 | 35.03% (197) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1700 | - | 1e-25 | 36.96% (184) | TetR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1711c | - | 7e-25 | 32.32% (198) | hypothetical protein Mb1711c |
| M. gilvum PYR-GCK | Mflv_0253 | - | 6e-29 | 37.56% (197) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1685c | - | 7e-25 | 32.32% (198) | hypothetical protein Rv1685c |
| M. leprae Br4923 | MLBr_02568 | - | 4e-07 | 43.14% (51) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2351c | - | 1e-22 | 34.67% (199) | TetR family transcriptional regulator |
| M. marinum M | MMAR_3397 | - | 2e-24 | 32.09% (187) | putative regulatory protein |
| M. avium 104 | MAV_3088 | - | 2e-24 | 33.50% (197) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3542 | - | 5e-23 | 36.31% (179) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_1664 | - | 2e-23 | 32.29% (192) | hypothetical protein MUL_1664 |
| M. vanbaalenii PYR-1 | Mvan_0641 | - | 7e-26 | 37.24% (196) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
TH_3542|M.thermoresistible__bu -----------------------VLAAAKRRFAQDGYEKTTLRAIARDAH
Mvan_0641|M.vanbaalenii_PYR-1 MPRPGETARRRGRRQGEPVSKEAVLAAAKTRFARDGYEKTTLRAIADDAR
Mflv_0253|M.gilvum_PYR-GCK MPPPAQTGRRRGRRKGDPVSRETVLAAAKGRFARDGFEKTTLRAIAEDAG
MSMEG_2182|M.smegmatis_MC2_155 ----MAENRRSGRWRTGQQNKQRILDAARDHFMRSGYEKATVRGIAAEAG
Mb1711c|M.bovis_AF2122/97 MAAPDNSRRRPGRPAGSSDTRERILSSARELFAHNGIDRTSIRAVAAKAG
Rv1685c|M.tuberculosis_H37Rv MAAPDNSRRRPGRPAGSSDTRERILSSARELFAHNGIDRTSIRAVAAKAG
MAV_3088|M.avium_104 MTTTGTTRRRRGRPAGSSGSRERILASARELFARNGIRNTSIRAVAAAAG
MUL_1664|M.ulcerans_Agy99 METTNTGRRRPGRPTGTSNTREHILTCARELFALNGLDRTSVRSVAAAAG
MAB_2351c|M.abscessus_ATCC_199 MSS-----KRPGRRPGTSSARDDILASARKLFSLNGIDKTSIRAIASDAG
MMAR_3397|M.marinum_M ----------MGRRRGNPDTRNAIVTTARRLFAESGYEQTSVRDIASAAG
MLBr_02568|M.leprae_Br4923 -------MAGGAKRIPRAVREQQMLDAAVQMFSVNGYHKTSMDAIAAEAQ
:: * * .* .::: :* *
TH_3542|M.thermoresistible__bu VDASMVLYLFGSKAELFRESLELILDPEMLVAAMT-----GDDGDVGTRM
Mvan_0641|M.vanbaalenii_PYR-1 VDASMVLYLFGSKEQLFREALRLILDPEVLVAALAGVE--GDSSDVGTRM
Mflv_0253|M.gilvum_PYR-GCK VDASMVLYLFGSKEALFRESLRLILDPGVLIEALS-----GGEADIGTRM
MSMEG_2182|M.smegmatis_MC2_155 VDVAMVYYFFGNKEGLFNAAVLDTPEHPLHQLAALLD---DGPEEIGARL
Mb1711c|M.bovis_AF2122/97 VDAALVHHYFGTKQQLFAAAIHIPIDPMVIIGPIREA----PVEELGYKL
Rv1685c|M.tuberculosis_H37Rv VDAALVHHYFGTKQQLFAAAIHIPIDPMVIIGPIREA----PVEELGYKL
MAV_3088|M.avium_104 VDSALVHHYFGTKEKLFAAAVQIPIDPMQVIGPLREV----PVDDLGYAL
MUL_1664|M.ulcerans_Agy99 VDASLVHHYYGTKQQLFAAAIQIPIDPTVVLEQIVET----PVDELGLKI
MAB_2351c|M.abscessus_ATCC_199 VDPALVHHYFGTKLDLFREVVQLPVDPSVVLQPLRDV----PVDELGVTI
MMAR_3397|M.marinum_M VDPALIRHYFGSKAELFRSIVGWPFEADAMSALILSG----DRDELGARL
MLBr_02568|M.leprae_Br4923 ISKPMLYLYYGSKEDLFGACLNREMSRFIAMVRADIDFTQSPKDLLRNTI
:. .:: :*.* ** : . : :
TH_3542|M.thermoresistible__bu VRVYLQIWESPDTGPTIRAMLQSATSNTDAREAFRSFLREYLLTAVSG-V
Mvan_0641|M.vanbaalenii_PYR-1 VRTYLGIWEAADSAASMRAMLASATSNPDAHEAFRSFMQDYVLTAVSG-V
Mflv_0253|M.gilvum_PYR-GCK VRAYLGVWESPESAESMRTMLASATSNPDAREAFRGFIQDYVLAAVVG-T
MSMEG_2182|M.smegmatis_MC2_155 VREVIRRWDEGGSFDLLLTVFKSADIHPLARKLLHDSLAGPVAQRVAA-E
Mb1711c|M.bovis_AF2122/97 PSLLLPIWDS-ELGAGLIATLRSLISGSD-VGLARSFLEEVVTVELGSRV
Rv1685c|M.tuberculosis_H37Rv PSLLLPIWDS-ELGAGLIATLRSLISGSD-VGLARSFLEEVVTVELGSRV
MAV_3088|M.avium_104 PSMLLPLWDS-EVGAAFIATLRSILAGEE-ISLFRTFIQDVIGVEVGPRV
MUL_1664|M.ulcerans_Agy99 PSVPLPLWDS-ELSTGLVATLRALMAGTE-SNLARSFLQEVVTSEVGSRV
MAB_2351c|M.abscessus_ATCC_199 PRLIIALWDS-ELGANMLAVFRSALTGAD-DGLVRVFFREVLVNIIAERV
MMAR_3397|M.marinum_M TRVFLEAWERPESRAPLLAILRGAATHEESTTLVRQFFQDQMYPQLAQPL
MLBr_02568|M.leprae_Br4923 VSFLRYIDANRASWIVMYIQAMSLPAFAQTVREARDQITELVAGLMRVGT
: . : : : :
TH_3542|M.thermoresistible__bu LGGGEQARLRAELAATGLVGTAMLRYIMRVPPLAELSTEEVVRMLAPTVN
Mvan_0641|M.vanbaalenii_PYR-1 LGGGEQARLRAMLAASSLVGTAMLRYVMRVAPLAELPTEDVVRLVAPTVT
Mflv_0253|M.gilvum_PYR-GCK LGGDDRARLRAMLAASSLVGTAMMRYLMEIPPLSELATEDVIRLVAPTVT
MSMEG_2182|M.smegmatis_MC2_155 FG-VDDAELRVELVTVHMVGLAFARYQLKIEPLASAGLDDLVSWLGPTVQ
Mb1711c|M.bovis_AF2122/97 DNPPGTGKIRTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNLQ
Rv1685c|M.tuberculosis_H37Rv DNPPGTGKIRTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNLQ
MAV_3088|M.avium_104 DNPAGSGVIRIQFVASQLVGVVMARYILELEPFASLPPEQIAATIAPNLQ
MUL_1664|M.ulcerans_Agy99 DSPTGTGRIRAQFVTSQLLGVVMARHIVKIEPFASLPADQVAQAIAPTLQ
MAB_2351c|M.abscessus_ATCC_199 DSPPGSGVLRAEFAITQMAGILVGRYIMAIEPLASLTAEQIALTVGPNIQ
MMAR_3397|M.marinum_M PGP--DTELRIDLAMAQLLGIAYLRHVLRLEPIASTPLEEVVARVTPTII
MLBr_02568|M.leprae_Br4923 RSSRPDHEYEIMAGALVGAGEAMATRLTTGDIAVDEAAELMINLFWHGLK
. . * . : : . :
TH_3542|M.thermoresistible__bu RYLTADADELGLPSSLTAGS
Mvan_0641|M.vanbaalenii_PYR-1 RYLTADARELGLPD------
Mflv_0253|M.gilvum_PYR-GCK RYLTAPADELGLPG------
MSMEG_2182|M.smegmatis_MC2_155 RYLTAPDP------------
Mb1711c|M.bovis_AF2122/97 RYLTGELPDDLAP-------
Rv1685c|M.tuberculosis_H37Rv RYLTGELPDDLAP-------
MAV_3088|M.avium_104 RYLTGDLPDGLAP-------
MUL_1664|M.ulcerans_Agy99 RYLNR---------------
MAB_2351c|M.abscessus_ATCC_199 RYLTGALPS-ITP-------
MMAR_3397|M.marinum_M AHLEPSRLDKAADR------
MLBr_02568|M.leprae_Br4923 GAPADRDFGPSIVAG-----