For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTITALTVTLPLLWRRLTTAGVKYADQGHFVGSAGVPAADAGGRDAASEQIARWTQTCTVVLVCGHGPAK WAFRSWCTSRSCDTLPVALRYRLQSNPLVGKLTTKYFLPLGTRQVGDHVVFFNFGYEEDPPMALPLSESD EPNRYCIQLYHQTASQVDLTGKEVLEVSCGAGGGASYIARNLGPASYTGLDLNPASIDLCRAKHRLPGLQ FVQGDAQNLPFPDESFDAVVNVEASHQYPDFRGFLAEVARVLRPGGHFLYTDSRRNPVVAEWEAALADAP LRTISQRDIGAQAKRGLDANTARSQEAIGRRAPVLLAGLTRCAVRVLDWDLRRGGGFSYRIYLFAKD
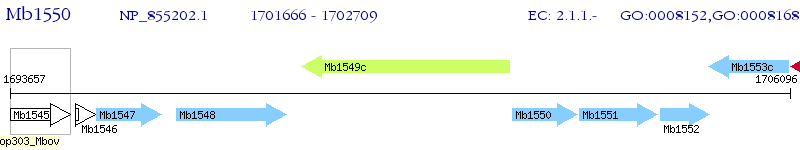
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1550 | - | - | 100% (347) | methyltransferase |
| M. bovis AF2122 / 97 | Mb2976 | - | 3e-79 | 59.76% (251) | methyltransferase (methylase) |
| M. bovis AF2122 / 97 | Mb3756 | - | 7e-11 | 32.16% (171) | transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2862 | - | 5e-12 | 35.71% (126) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv1523 | - | 0.0 | 100.00% (347) | methyltransferase |
| M. leprae Br4923 | MLBr_00130 | - | 1e-76 | 55.08% (256) | hypothetical protein MLBr_00130 |
| M. abscessus ATCC 19977 | MAB_4103c | - | 3e-78 | 58.04% (255) | methyltransferase |
| M. marinum M | MMAR_1766 | - | 2e-76 | 57.31% (253) | methyltransferase |
| M. avium 104 | MAV_3877 | - | 4e-10 | 36.79% (106) | hypothetical protein MAV_3877 |
| M. smegmatis MC2 155 | MSMEG_0393 | - | 1e-72 | 53.82% (262) | Fmt protein |
| M. thermoresistible (build 8) | TH_1636 | - | 3e-77 | 62.07% (232) | POSSIBLE METHYLTRANSFERASE (METHYLASE) |
| M. ulcerans Agy99 | MUL_2377 | - | 7e-75 | 57.26% (248) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_3244 | - | 1e-11 | 30.54% (167) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2862|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3244|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb1550|M.bovis_AF2122/97 MTITALTVTLPLLWRRLTTAGVKYADQGHFVGSAGVPAADAGGRDAASEQ
Rv1523|M.tuberculosis_H37Rv MTITALTVTLPLLWRRLTTAGVKYADQGHFVGSAGVPAADAGGRDAASEQ
MSMEG_0393|M.smegmatis_MC2_155 --------------------------------------------------
TH_1636|M.thermoresistible__bu --------------------------------------------------
MAB_4103c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00130|M.leprae_Br4923 --------------------------------------------------
MUL_2377|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1766|M.marinum_M --------------------------------------------------
MAV_3877|M.avium_104 --------------------------------------------------
Mflv_2862|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3244|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb1550|M.bovis_AF2122/97 IARWTQTCTVVLVCGHGPAKWAFRSWCTSRSCDTLPVALRYRLQSNPLVG
Rv1523|M.tuberculosis_H37Rv IARWTQTCTVVLVCGHGPAKWAFRSWCTSRSCDTLPVALRYRLQSNPLVG
MSMEG_0393|M.smegmatis_MC2_155 ----------------------------MEARDRVAWRL------NIKVA
TH_1636|M.thermoresistible__bu ---------------------------VRESRDQVFRKL------NEKLT
MAB_4103c|M.abscessus_ATCC_199 ---------------------------MQRSRDWVMFKL------NAVFT
MLBr_00130|M.leprae_Br4923 -------------------------MAFTRIHSFLASAG------NTSMY
MUL_2377|M.ulcerans_Agy99 -------------------------MAFSPAHRLLARLG------STSIY
MMAR_1766|M.marinum_M -------------------------MALSRTHRVVAGVA------HTRVY
MAV_3877|M.avium_104 ---------------------------MSALVPDLP--------PHPPAG
Mflv_2862|M.gilvum_PYR-GCK ------MNMAETALINSAPRRWLQRVY--EVPVLLRFGGRLVP-------
Mvan_3244|M.vanbaalenii_PYR-1 ------MNRAETALINSPPRRWLQRLY--EVPVLLRFGGRLPP-------
Mb1550|M.bovis_AF2122/97 KLTTKYFLPLGTRQVG-DHVVFFNFGYEEDPPMALPLSESDEPNRYCIQL
Rv1523|M.tuberculosis_H37Rv KLTTKYFLPLGTRQVG-DHVVFFNFGYEEDPPMALPLSESDEPNRYCIQL
MSMEG_0393|M.smegmatis_MC2_155 QQAQKLVYRYATRRLKDDDVVFLNYGYEEDPPMGIPLSESDELNRYSIQL
TH_1636|M.thermoresistible__bu QHGQKFIYSYASRRLDDDDVVFLNYGYEEDPPMGLPLSETDEPNRYSIQL
MAB_4103c|M.abscessus_ATCC_199 QRVQKFIYRHATRKLETEDVVFLNYGYEEEPAMGVPLSASDEPDRYSIQL
MLBr_00130|M.leprae_Br4923 KRVWRFWYPLMTHKLGTDEIMFINWAYEEDPPMALPLEASDEPNRAHINL
MUL_2377|M.ulcerans_Agy99 KRVWRYWYPLMTRGLGADKIAFLNWAYEEDPPIDLTLEVSDEPNRDHINM
MMAR_1766|M.marinum_M KKIWKYWYPLMTRGLGADELVFINWAYEEDPPMDLPLEATDEPDRCHINL
MAV_3877|M.avium_104 AAPAGDAVMMLTGERTIPGLDIENYWFRRHEVVYQRLAR-----------
: : : . : :
Mflv_2862|M.gilvum_PYR-GCK -----------DTQALEIGCGSGYGTQLILERFGAATVDAVDLDPAMIRR
Mvan_3244|M.vanbaalenii_PYR-1 -----------GSGALEIGCGSGYGSQLVLQRFGAARIDAVDLDPAMIER
Mb1550|M.bovis_AF2122/97 YHQTASQVDLTGKEVLEVSCGAGGGASYIARNLGPASYTGLDLNPASIDL
Rv1523|M.tuberculosis_H37Rv YHQTASQVDLTGKEVLEVSCGAGGGASYIARNLGPASYTGLDLNPASIDL
MSMEG_0393|M.smegmatis_MC2_155 YHSTAAQADVEGKRVLEVGCGHGGGASYLARTFRPATYTGLDLNSDGINF
TH_1636|M.thermoresistible__bu YHRTAGQADLTGKRVLEVGCGHGGGASYLMRTLGPMSYVGLDLNPAGIEF
MAB_4103c|M.abscessus_ATCC_199 YHSTATQADLGGQRVLEVGCGHGGGASYLVRALQPASYTGLDLNPDGISF
MLBr_00130|M.leprae_Br4923 YHRTATQVNLSGKRILEVSCGHGGGASYLTRALHPASYTGLDLNPAGIKL
MUL_2377|M.ulcerans_Agy99 YHRTATHVELSGKRVLEVSCGHGGGASYLTRTLHPASYTGLDLNRAGIKL
MMAR_1766|M.marinum_M YHRTATQADLSGKRVLEVSCGHGGGASYLTRTLGPASYTALDLNPAGIKF
MAV_3877|M.avium_104 --------HCAGRDVLEAGCGEGYGADLIAG--VARRVIAVDYDEAAVAH
. ** .** * *:. : . .:* : :
Mflv_2862|M.gilvum_PYR-GCK AGERLARYGNRVRLVQGSATDLSTALDAGDGSYGAVFDFGIVHHIPDWRN
Mvan_3244|M.vanbaalenii_PYR-1 ARTRLAPYGDRVHLATGSATDLREALNADDGSYDAVFDFGIIHHIPDWRA
Mb1550|M.bovis_AF2122/97 CRAKHRLPG--LQFVQGDAQNLP----FPDESFDAVVNVEASHQYPDFRG
Rv1523|M.tuberculosis_H37Rv CRAKHRLPG--LQFVQGDAQNLP----FPDESFDAVVNVEASHQYPDFRG
MSMEG_0393|M.smegmatis_MC2_155 CRRRHNIAG--LEFVQGDAQDLP----FPDKNFDAVLNVESSHLYPRFDV
TH_1636|M.thermoresistible__bu CRKKHRLPG--LEFVVGDAQDLP----FGAESFDAVINIESSHLYPRFSR
MAB_4103c|M.abscessus_ATCC_199 CRRRHDLPG--LEFVQGDAEDLP----FPDESFDAVINVESSHLYPHFPV
MLBr_00130|M.leprae_Br4923 CQKRHQLPG--LEFVRGDAENLP----FDNESFDVVINIEASHCYPHFPR
MUL_2377|M.ulcerans_Agy99 CQRRHNLPG--LDFVRGDAENLP----FEDESFDVVLKVEASHCYPHFSR
MMAR_1766|M.marinum_M CQQRHHLPG--LDFVQGDAEDLP----FEDESFDVVLNVEASHCYPRFPV
MAV_3877|M.avium_104 VRGRYPRVD----VMQANLAQLP----LPDSSVDVVVNFQVIEHLWDQTQ
: . . .. :* . ..*... .
Mflv_2862|M.gilvum_PYR-GCK AVAEVARVLAPGGRFYFDEVTAHALARPSYKRLFDHPT---EDRFSAEQF
Mvan_3244|M.vanbaalenii_PYR-1 AVAEAARVLIPGGRFYFEEVTAHALSRPTYRLLFDHPT---DDRFTAEDF
Mb1550|M.bovis_AF2122/97 FLAEVARVLRPGGHFLYTDSRRNPVVAEWEAALADAPLRTISQRDIGAQA
Rv1523|M.tuberculosis_H37Rv FLAEVARVLRPGGHFLYTDSRRNPVVAEWEAALADAPLRTISQRDIGAQA
MSMEG_0393|M.smegmatis_MC2_155 FLTEVARVLRPGGYFLYTDARPRYDIPEWERALADAPLQMLSQRAINFEV
TH_1636|M.thermoresistible__bu FLSEVARVLRPGGHFLYADARQPHEFAEWEAALAAAPLDMVSARVINAEV
MAB_4103c|M.abscessus_ATCC_199 FLTEVARVLRPGGNFLYTDARSVQDVAGWKVALANAPLRMVSERGINAEV
MLBr_00130|M.leprae_Br4923 FLAEVVRVLRPGGHLAYADLRPSNKVGEWEVDFANSRLQQLSQREINAEV
MUL_2377|M.ulcerans_Agy99 FLAEVVRVLRPGGYLLYTDLRPSNEIAEWEADLAGSPLRQLSQREINAEV
MMAR_1766|M.marinum_M FLEEVKRVLRPGGYFAYADIRPCTEIAEWEAALAAAGLQQISHREINAEV
MAV_3877|M.avium_104 FVVECARVLRPSGLLLMSTPNRITFSPGRDTPINPFHTRELNAVELTELL
: * *** *.* : : .
Mflv_2862|M.gilvum_PYR-GCK SDE------LA--HHGLVVLGSRTRIQGDYLLGVAAKP----------LP
Mvan_3244|M.vanbaalenii_PYR-1 LDE------LG--RHGLVILGSVTRIQGDYLLGVAAKP----------LA
Mb1550|M.bovis_AF2122/97 KRG------LD--ANTARSQEAIGRRAPVLLAGLTR---CAVRVLDWDLR
Rv1523|M.tuberculosis_H37Rv KRG------LD--ANTARSQEAIGRRAPVLLAGLTR---CAVRVLDWDLR
MSMEG_0393|M.smegmatis_MC2_155 VRG------ME--KNLDALESVVDRVAPALLRDWIQKY-GPARRAYEELR
TH_1636|M.thermoresistible__bu VRG------ME--QILPVWEDVIERVAPAFLRRRVREF-APARRAWEELR
MAB_4103c|M.abscessus_ATCC_199 RRG------ME--KNLARWRYVIDRATPAPLRGIVRRF-APAQRAYDDLR
MLBr_00130|M.leprae_Br4923 LRG------IA--SNSQKSRDLVDRHLPAFLRFAGREFIGVQGTQLSRYL
MUL_2377|M.ulcerans_Agy99 VRG------IE--KNSHTSRVLVDRNLPAFLRFADR----AVGRQLSRYL
MMAR_1766|M.marinum_M LRG------ID--INTPKSRELVKRHLPIFLRAAGRNYIGATGTPLYRLM
MAV_3877|M.avium_104 VGGGFRDVSISGLFHGPRLREMDARHGGSIIDAQIARAVADAPWPPQLAA
: * :
Mflv_2862|M.gilvum_PYR-GCK TGCE----------------------------------
Mvan_3244|M.vanbaalenii_PYR-1 ADGGR---------------------------------
Mb1550|M.bovis_AF2122/97 RGGGFSYRIYLFAKD-----------------------
Rv1523|M.tuberculosis_H37Rv RGGGFSYRIYLFAKD-----------------------
MSMEG_0393|M.smegmatis_MC2_155 -ENTTEYRMYCFVKPAE---------------------
TH_1636|M.thermoresistible__bu TEGSTEYRMYCFTKPPEHADIP----------------
MAB_4103c|M.abscessus_ATCC_199 AGGSVEYRMYRFVKA-----------------------
MLBr_00130|M.leprae_Br4923 EGGELSYRMYSFAKD-----------------------
MUL_2377|M.ulcerans_Agy99 EGGELSYRMYCFTKDFAASR------------------
MMAR_1766|M.marinum_M QSGDISYRMYCFVKE-----------------------
MAV_3877|M.avium_104 DVAAVTVDDFELVPSGADAASGHHIDDSLDLIAIAVAP