For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QRSRDWVMFKLNAVFTQRVQKFIYRHATRKLETEDVVFLNYGYEEEPAMGVPLSASDEPDRYSIQLYHST ATQADLGGQRVLEVGCGHGGGASYLVRALQPASYTGLDLNPDGISFCRRRHDLPGLEFVQGDAEDLPFPD ESFDAVINVESSHLYPHFPVFLTEVARVLRPGGNFLYTDARSVQDVAGWKVALANAPLRMVSERGINAEV RRGMEKNLARWRYVIDRATPAPLRGIVRRFAPAQRAYDDLRAGGSVEYRMYRFVKA*
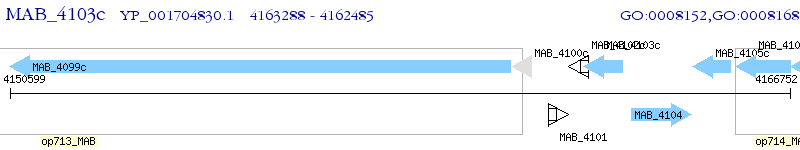
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4103c | - | - | 100% (267) | methyltransferase |
| M. abscessus ATCC 19977 | MAB_0018c | - | 7e-12 | 30.00% (180) | putative methyltransferase |
| M. abscessus ATCC 19977 | MAB_3364 | - | 2e-11 | 34.51% (142) | hypothetical protein MAB_3364 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1550 | - | 2e-78 | 58.04% (255) | methyltransferase |
| M. gilvum PYR-GCK | Mflv_4060 | - | 3e-11 | 39.80% (98) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv1523 | - | 2e-78 | 58.04% (255) | methyltransferase |
| M. leprae Br4923 | MLBr_00130 | - | 1e-79 | 56.47% (255) | hypothetical protein MLBr_00130 |
| M. marinum M | MMAR_1766 | - | 1e-78 | 58.00% (250) | methyltransferase |
| M. avium 104 | MAV_3903 | - | 4e-10 | 34.91% (106) | methyltransferase, UbiE/COQ5 family protein |
| M. smegmatis MC2 155 | MSMEG_0393 | - | 1e-102 | 67.68% (263) | Fmt protein |
| M. thermoresistible (build 8) | TH_1636 | - | 1e-101 | 68.80% (266) | POSSIBLE METHYLTRANSFERASE (METHYLASE) |
| M. ulcerans Agy99 | MUL_2009 | - | 2e-75 | 59.91% (227) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_0109 | - | 4e-12 | 39.58% (96) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0393|M.smegmatis_MC2_155 --------------------------------------------------
TH_1636|M.thermoresistible__bu --------------------------------------------------
MAB_4103c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1550|M.bovis_AF2122/97 MTITALTVTLPLLWRRLTTAGVKYADQGHFVGSAGVPAADAGGRDAASEQ
Rv1523|M.tuberculosis_H37Rv MTITALTVTLPLLWRRLTTAGVKYADQGHFVGSAGVPAADAGGRDAASEQ
MMAR_1766|M.marinum_M --------------------------------------------------
MUL_2009|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00130|M.leprae_Br4923 --------------------------------------------------
Mvan_0109|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4060|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3903|M.avium_104 --------------------------------------------------
MSMEG_0393|M.smegmatis_MC2_155 ----------------------------MEARD------RVAWRLNIKVA
TH_1636|M.thermoresistible__bu ---------------------------VRESRD------QVFRKLNEKLT
MAB_4103c|M.abscessus_ATCC_199 ---------------------------MQRSRD------WVMFKLNAVFT
Mb1550|M.bovis_AF2122/97 IARWTQTCTVVLVCGHGPAKWAFRSWCTSRSCDTLPVALRYRLQSNPLVG
Rv1523|M.tuberculosis_H37Rv IARWTQTCTVVLVCGHGPAKWAFRSWCTSRSCDTLPVALRYRLQSNPLVG
MMAR_1766|M.marinum_M -------------------------MALSRTHR------VVAGVAHTRVY
MUL_2009|M.ulcerans_Agy99 -------------------------MALSRTHR------VVAGVAHTRVY
MLBr_00130|M.leprae_Br4923 -------------------------MAFTRIHS------FLASAGNTSMY
Mvan_0109|M.vanbaalenii_PYR-1 -----------------------MTHTVLRMTER-------LLIDPPEHP
Mflv_4060|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3903|M.avium_104 ----------------------------------------MTRSTDAVPT
MSMEG_0393|M.smegmatis_MC2_155 QQAQKLVYRYATRRLKDDDVVFLNYGYEEDPPMGIPLSES--DELNRYSI
TH_1636|M.thermoresistible__bu QHGQKFIYSYASRRLDDDDVVFLNYGYEEDPPMGLPLSET--DEPNRYSI
MAB_4103c|M.abscessus_ATCC_199 QRVQKFIYRHATRKLETEDVVFLNYGYEEEPAMGVPLSAS--DEPDRYSI
Mb1550|M.bovis_AF2122/97 KLTTKYFLPLGTRQVG-DHVVFFNFGYEEDPPMALPLSES--DEPNRYCI
Rv1523|M.tuberculosis_H37Rv KLTTKYFLPLGTRQVG-DHVVFFNFGYEEDPPMALPLSES--DEPNRYCI
MMAR_1766|M.marinum_M KKIWKYWYPLMTRGLGADELVFINWAYEEDPPMDLPLEAT--DEPDRCHI
MUL_2009|M.ulcerans_Agy99 KKIWKYWYPLMTRGLGADELVFINWAYEEDPPMDLPLEAT--DEPDRCHI
MLBr_00130|M.leprae_Br4923 KRVWRFWYPLMTHKLGTDEIMFINWAYEEDPPMALPLEAS--DEPNRAHI
Mvan_0109|M.vanbaalenii_PYR-1 DTSKGYLDLLGDEPDSNRGAIQALWASRLG---SMFYDNA--QTLARRLM
Mflv_4060|M.gilvum_PYR-GCK -----MATVDNEDDRALKAKHRALWASGDYPAVAAELIPS--LGP-----
MAV_3903|M.avium_104 PHATAEQVEAARHDSKLAQVLYHDWEAETYDEKWSISYDQRCIDYARGRF
:
MSMEG_0393|M.smegmatis_MC2_155 QLYHSTAAQADVE-GKRVLEVGCGHGGGASYLART-FRP-ATYTGLDLNS
TH_1636|M.thermoresistible__bu QLYHRTAGQADLT-GKRVLEVGCGHGGGASYLMRT-LGP-MSYVGLDLNP
MAB_4103c|M.abscessus_ATCC_199 QLYHSTATQADLG-GQRVLEVGCGHGGGASYLVRA-LQP-ASYTGLDLNP
Mb1550|M.bovis_AF2122/97 QLYHQTASQVDLT-GKEVLEVSCGAGGGASYIARN-LGP-ASYTGLDLNP
Rv1523|M.tuberculosis_H37Rv QLYHQTASQVDLT-GKEVLEVSCGAGGGASYIARN-LGP-ASYTGLDLNP
MMAR_1766|M.marinum_M NLYHRTATQADLS-GKRVLEVSCGHGGGASYLTRT-LGP-ASYTALDLNP
MUL_2009|M.ulcerans_Agy99 NLYHRTATQADLS-GKRVLEVSCGHGGGASYLTRT-LGP-ASYTALDLNP
MLBr_00130|M.leprae_Br4923 NLYHRTATQVNLS-GKRILEVSCGHGGGASYLTRA-LHP-ASYTGLDLNP
Mvan_0109|M.vanbaalenii_PYR-1 TAWQQPTEWLNVPVGGTALDVGSGPGNVTGALGRA-VGPGGLALGVDISE
Mflv_4060|M.gilvum_PYR-GCK ----ELVRACGVRSGDRVLDVAAGSGN-ASIPAAA-AGAVVTASDLTPEL
MAV_3903|M.avium_104 DAIVPEHVLRELP-YDRALELGCGTGFFLLNLIQSGVARRGSVTDLSPGM
: *::..* * :
MSMEG_0393|M.smegmatis_MC2_155 DGINFCRRRHNIAGLEFVQGDAQDLPFPDKNFDAVLNVESSHLYPRFDVF
TH_1636|M.thermoresistible__bu AGIEFCRKKHRLPGLEFVVGDAQDLPFGAESFDAVINIESSHLYPRFSRF
MAB_4103c|M.abscessus_ATCC_199 DGISFCRRRHDLPGLEFVQGDAEDLPFPDESFDAVINVESSHLYPHFPVF
Mb1550|M.bovis_AF2122/97 ASIDLCRAKHRLPGLQFVQGDAQNLPFPDESFDAVVNVEASHQYPDFRGF
Rv1523|M.tuberculosis_H37Rv ASIDLCRAKHRLPGLQFVQGDAQNLPFPDESFDAVVNVEASHQYPDFRGF
MMAR_1766|M.marinum_M AGIKFCQQRHHLPGLDFVQGDAEDLPFEDESFDVVLNVEASHCYPRFPVF
MUL_2009|M.ulcerans_Agy99 AGIKFCQQRHHLPGLDFVQGDAEDLPFEDESFDVVLNVEASHCYPRFPVF
MLBr_00130|M.leprae_Br4923 AGIKLCQKRHQLPGLEFVRGDAENLPFDNESFDVVINIEASHCYPHFPRF
Mvan_0109|M.vanbaalenii_PYR-1 PMLARAVRAEAGPNVGFIRADAQHLPFRDESFDAVVSIAMLQLIPDPAVA
Mflv_4060|M.gilvum_PYR-GCK FDAGRRIAAERGVAVEWVEADAEALPFPDDSFDVVMSCVGAMFAPHHQAT
MAV_3903|M.avium_104 VKVATRNGQSLGLDIDGRVADAEGIPYEDDTFDLVVGHAVLHHIPDVELS
: .**: :*: ..** *:. *
MSMEG_0393|M.smegmatis_MC2_155 LTEVARVLRPGGYFLYTDARPRY--------DIPEWERALADAPLQMLSQ
TH_1636|M.thermoresistible__bu LSEVARVLRPGGHFLYADARQPH--------EFAEWEAALAAAPLDMVSA
MAB_4103c|M.abscessus_ATCC_199 LTEVARVLRPGGNFLYTDARSVQ--------DVAGWKVALANAPLRMVSE
Mb1550|M.bovis_AF2122/97 LAEVARVLRPGGHFLYTDSRRNP--------VVAEWEAALADAPLRTISQ
Rv1523|M.tuberculosis_H37Rv LAEVARVLRPGGHFLYTDSRRNP--------VVAEWEAALADAPLRTISQ
MMAR_1766|M.marinum_M LEEVKRVLRPGGYFAYADIRPCT--------EIAEWEAALAAAGLQQISH
MUL_2009|M.ulcerans_Agy99 LEEVKRVLRPGGYFAYADIRPCT--------EIAEWEAALAAAGLQQISH
MLBr_00130|M.leprae_Br4923 LAEVVRVLRPGGHLAYADLRPSN--------KVGEWEVDFANSRLQQLSQ
Mvan_0109|M.vanbaalenii_PYR-1 LAEMVRVLRPGRRMAVMVP-----------------TAGPAAGLLRYLPN
Mflv_4060|M.gilvum_PYR-GCK ADEIVRVTRPGGTLGLISWTPEG--------FIGHLFATMKPYAPPPPPG
MAV_3903|M.avium_104 LREVIRVLRPGGRFVFAGEPTSAGDVYARELSTLTWRIATNVTKLPGLGS
*: ** *** :
MSMEG_0393|M.smegmatis_MC2_155 R-------------AINFEVVRGMEKNLDALESVVDRVAPALLRDWIQKY
TH_1636|M.thermoresistible__bu R-------------VINAEVVRGMEQILPVWEDVIERVAPAFLRRRVREF
MAB_4103c|M.abscessus_ATCC_199 R-------------GINAEVRRGMEKNLARWRYVIDRATPAPLRGIVRRF
Mb1550|M.bovis_AF2122/97 R-------------DIGAQAKRGLDANTARSQEAIGRRAPVLLAGLTR--
Rv1523|M.tuberculosis_H37Rv R-------------DIGAQAKRGLDANTARSQEAIGRRAPVLLAGLTR--
MMAR_1766|M.marinum_M R-------------EINAEVLRGIDINTPKSRELVKRHLPIFLRAAGRNY
MUL_2009|M.ulcerans_Agy99 R-------------EINAEVLRGIDINTPKSRERVKRHLPIFLRAAGRNY
MLBr_00130|M.leprae_Br4923 R-------------EINAEVLRGIASNSQKSRDLVDRHLPAFLRFAGREF
Mvan_0109|M.vanbaalenii_PYR-1 A-------------GAHAFGDDELGDTFESLGLAGVRINTVGTIQWVRGR
Mflv_4060|M.gilvum_PYR-GCK ASPPPMWGSENHVEALFGDRVRDLTMRRQTVRFDHCASPLDFREYWKRNY
MAV_3903|M.avium_104 WR------------RPQAELDESSRAAALEAIVDLHTFTPGDLERMAANA
MSMEG_0393|M.smegmatis_MC2_155 GPAR-RAYEELR-ENTTEYRMYCFVKPAE---------------------
TH_1636|M.thermoresistible__bu APAR-RAWEELRTEGSTEYRMYCFTKPPEHADIP----------------
MAB_4103c|M.abscessus_ATCC_199 APAQ-RAYDDLRAGGSVEYRMYRFVKA-----------------------
Mb1550|M.bovis_AF2122/97 CAVR-VLDWDLRRGGGFSYRIYLFAKD-----------------------
Rv1523|M.tuberculosis_H37Rv CAVR-VLDWDLRRGGGFSYRIYLFAKD-----------------------
MMAR_1766|M.marinum_M IGATGTPLYRLMQSGDISYRMYCFVKE-----------------------
MUL_2009|M.ulcerans_Agy99 IGATGTPPIPPDAKR-----------------------------------
MLBr_00130|M.leprae_Br4923 IGVQGTQLSRYLEGGELSYRMYSFAKD-----------------------
Mvan_0109|M.vanbaalenii_PYR-1 KS------------------------------------------------
Mflv_4060|M.gilvum_PYR-GCK GPTLAAYRFNEDRPERLAELDAAFLDFLTTWNQADGTDGTAYDAEYLLVT
MAV_3903|M.avium_104 GATEVRTVSEEFTAAMFGWPVRTFEASVPPGRLGWGWAKFAFNGWKTLSW
MSMEG_0393|M.smegmatis_MC2_155 --------------------------
TH_1636|M.thermoresistible__bu --------------------------
MAB_4103c|M.abscessus_ATCC_199 --------------------------
Mb1550|M.bovis_AF2122/97 --------------------------
Rv1523|M.tuberculosis_H37Rv --------------------------
MMAR_1766|M.marinum_M --------------------------
MUL_2009|M.ulcerans_Agy99 --------------------------
MLBr_00130|M.leprae_Br4923 --------------------------
Mvan_0109|M.vanbaalenii_PYR-1 --------------------------
Mflv_4060|M.gilvum_PYR-GCK ATKR----------------------
MAV_3903|M.avium_104 VDANIWRRVVPKGWFYNVMVTGVKPT