For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNRAETALINSPPRRWLQRLYEVPVLLRFGGRLPPGSGALEIGCGSGYGSQLVLQRFGAARIDAVDLDPA MIERARTRLAPYGDRVHLATGSATDLREALNADDGSYDAVFDFGIIHHIPDWRAAVAEAARVLIPGGRFY FEEVTAHALSRPTYRLLFDHPTDDRFTAEDFLDELGRHGLVILGSVTRIQGDYLLGVAAKPLAADGGR
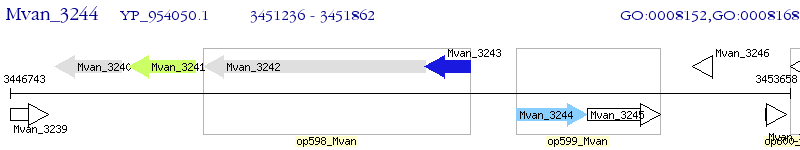
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3244 | - | - | 100% (208) | methyltransferase type 11 |
| M. vanbaalenii PYR-1 | Mvan_2090 | - | 8e-12 | 34.58% (107) | methyltransferase type 11 |
| M. vanbaalenii PYR-1 | Mvan_5517 | - | 1e-11 | 41.12% (107) | methyltransferase type 11 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3064c | - | 1e-12 | 34.62% (130) | hypothetical protein Mb3064c |
| M. gilvum PYR-GCK | Mflv_2862 | - | 5e-91 | 79.21% (202) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv3038c | - | 1e-12 | 34.62% (130) | hypothetical protein Rv3038c |
| M. leprae Br4923 | MLBr_01723 | - | 1e-12 | 35.51% (107) | hypothetical protein MLBr_01723 |
| M. abscessus ATCC 19977 | MAB_3381c | - | 2e-12 | 37.38% (107) | hypothetical protein MAB_3381c |
| M. marinum M | MMAR_0694 | - | 1e-14 | 35.44% (158) | hypothetical protein MMAR_0694 |
| M. avium 104 | MAV_3903 | - | 2e-12 | 33.33% (129) | methyltransferase, UbiE/COQ5 family protein |
| M. smegmatis MC2 155 | MSMEG_2329 | - | 1e-11 | 33.64% (107) | methyltransferase, UbiE/COQ5 family protein |
| M. thermoresistible (build 8) | TH_2189 | - | 1e-12 | 37.38% (107) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1900 | - | 2e-12 | 34.88% (129) | hypothetical protein MUL_1900 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3064c|M.bovis_AF2122/97 --------------------------------------------------
Rv3038c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1900|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2329|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu VIRRLMATALTALTTAVLAGCGDAGSWVEAKPAEGWSAQYGDAANSSYTS
MAV_3903|M.avium_104 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
MAB_3381c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3244|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2862|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0694|M.marinum_M --------------------------------------------------
Mb3064c|M.bovis_AF2122/97 --------------------------------------------------
Rv3038c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1900|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2329|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu AAGAEALTLEWSRSVKGELAAQVAVGASGYLAVNAQTPAGCSLMVWEYAN
MAV_3903|M.avium_104 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
MAB_3381c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3244|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2862|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0694|M.marinum_M --------------------------------------------------
Mb3064c|M.bovis_AF2122/97 --------------------------------------------------
Rv3038c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1900|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2329|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu SARQRWCTRLVQGGGRTSPLLDGFDNVYIGQPGAILSFPPTQWIRWRKPV
MAV_3903|M.avium_104 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
MAB_3381c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3244|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2862|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0694|M.marinum_M --------------------------------------------------
Mb3064c|M.bovis_AF2122/97 --------------------------------------------------
Rv3038c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1900|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2329|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu IGMPTTPRILAPGELLVVTHLGQVLLFDAHRGTVTGTPLDLVAGVDPTDS
MAV_3903|M.avium_104 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
MAB_3381c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3244|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2862|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0694|M.marinum_M --------------------------------------------------
Mb3064c|M.bovis_AF2122/97 ------------------------------------------MTRSSNIP
Rv3038c|M.tuberculosis_H37Rv ------------------------------------------MTRSSNIP
MUL_1900|M.ulcerans_Agy99 ------------------------------------------MTRSSEIP
MSMEG_2329|M.smegmatis_MC2_155 ------------------------------------------MTEIRDAA
TH_2189|M.thermoresistible__bu ERGLADCAGARXXXCWPPPRPPSTVTPRARDAARHPTARLPAMTETTEPG
MAV_3903|M.avium_104 ------------------------------------------MTRSTDAV
MLBr_01723|M.leprae_Br4923 ------------------------------------------MTRSTEIS
MAB_3381c|M.abscessus_ATCC_199 ------------------------------------------MTEITGLD
Mvan_3244|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2862|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0694|M.marinum_M ----------------------------MLNALSPDAQQESGAACAIPSS
Mb3064c|M.bovis_AF2122/97 AD-------ATPNPHATAEQVAAARHDSKLAQVLYHDWEAENYDEKWSIS
Rv3038c|M.tuberculosis_H37Rv AD-------ATPNPHATAEQVAAARHDSKLAQVLYHDWEAENYDEKWSIS
MUL_1900|M.ulcerans_Agy99 AD-------AAPNPHATAEEVEAARHDSKLAQVLYHDWEAENYDEKWSIS
MSMEG_2329|M.smegmatis_MC2_155 D--------PAPNPHATAEEVEAAMHDSKLAQVLYHDWEAETYDEKWSIS
TH_2189|M.thermoresistible__bu IGSEGAGDLPAPNPHATAEQVEAAMRDSKLAQILYHDWEAESYDDKWSIS
MAV_3903|M.avium_104 P-----------TPHATAEQVEAARHDSKLAQVLYHDWEAETYDEKWSIS
MLBr_01723|M.leprae_Br4923 AN-------AVPNPHATAEQVAAARKDSKLAQVLYHDWEAENYDEKWSIS
MAB_3381c|M.abscessus_ATCC_199 GQ---------PTTKATAEQVAAAFEDNKLAQILYHDWEAETYDEKWSIS
Mvan_3244|M.vanbaalenii_PYR-1 ----------------------MNRAETALINSPPRRWLQRLYEVPVLLR
Mflv_2862|M.gilvum_PYR-GCK ----------------------MNMAETALINSAPRRWLQRVYEVPVLLR
MMAR_0694|M.marinum_M GGAQPQSSTESGGPASHDYTDAMTRDFYDHEDSTYRGFWDPDGSLHWGVF
: : : . :
Mb3064c|M.bovis_AF2122/97 YDQRCVDYARGRFDAIVPDEVIAQLPYDRALELGCGTGFFLLNLIQAGVA
Rv3038c|M.tuberculosis_H37Rv YDQRCVDYARGRFDAIVPDEVIAQLPYDRALELGCGTGFFLLNLIQAGVA
MUL_1900|M.ulcerans_Agy99 YDQRCVEYARGRFDAIVPAQVHADLPYERALELGCGTGFFLLNLIQAGVA
MSMEG_2329|M.smegmatis_MC2_155 YDQRCIDYARGRFDAIVPEDEQRKLPYDRALELGCGTGFFLLNLIQSGVA
TH_2189|M.thermoresistible__bu YDQRCVDYARGRFDAIVPDHVQRELPYDNALELGCGTGFFLLNLIQSGVA
MAV_3903|M.avium_104 YDQRCIDYARGRFDAIVPEHVLRELPYDRALELGCGTGFFLLNLIQSGVA
MLBr_01723|M.leprae_Br4923 YDQRCVDYVRHCFDAIVPDELFTELPYDCALELGCGTGFFLLNLIQSGVA
MAB_3381c|M.abscessus_ATCC_199 YDQRCIDYARGRFDAIVPVEDQRELPYDRALELGCGTGFFLLNLMQAGVA
Mvan_3244|M.vanbaalenii_PYR-1 FGGRLP-------------------PGSGALEIGCGSGYGSQLVLQRFGA
Mflv_2862|M.gilvum_PYR-GCK FGGRLV-------------------PDTQALEIGCGSGYGTQLILERFGA
MMAR_0694|M.marinum_M EDDDTGFIAGCQHWNEQMLRAARIGPQSRVLDLGCGNG-TVSAWLARQTG
. * .*::***.* : .
Mb3064c|M.bovis_AF2122/97 RRGSVTDLSPGMVKVA----TRNGQALGLDIDGRVADAEGIPYDDDAFDL
Rv3038c|M.tuberculosis_H37Rv RRGSVTDLSPGMVKVA----TRNGQALGLDIDGRVADAEGIPYDDDAFDL
MUL_1900|M.ulcerans_Agy99 RRGSVTDLSPGMVKVA----TRNGQSLGLDIDGRVADAEGIPYDDDTFDL
MSMEG_2329|M.smegmatis_MC2_155 RRGSVTDLSPGMVKVA----TRNGQSLGLDIDGRVADAEGIPYEDDTFDL
TH_2189|M.thermoresistible__bu RRGSVTDLSPGMVRVA----TRNGRSLGLDVDGRVADAEGIPYDDNTFDL
MAV_3903|M.avium_104 RRGSVTDLSPGMVKVA----TRNGQSLGLDIDGRVADAEGIPYEDDTFDL
MLBr_01723|M.leprae_Br4923 RRGSVTDLSPGMVKIA----TRNGQSLGLDINGRVADAEGIPYDDNTFDL
MAB_3381c|M.abscessus_ATCC_199 RRGSVTDLSPGMVKVA----TRTGQELGLDVDGRVADAERIPYDDNTFDL
Mvan_3244|M.vanbaalenii_PYR-1 ARIDAVDLDPAMIERARTRLAPYGDRVHLATGSATDLREALNADDGSYDA
Mflv_2862|M.gilvum_PYR-GCK ATVDAVDLDPAMIRRAGERLARYGNRVRLVQGSATDLSTALDAGDGSYGA
MMAR_0694|M.marinum_M AQVTGIDLSAVRIANAQ---RLAADNPQLQLEFMASSASALPFADGTFTH
**.. : * . * . : *.::
Mb3064c|M.bovis_AF2122/97 VVGHAVLHHIPDVELSLREVVRVLKPGGRFVFAG----------------
Rv3038c|M.tuberculosis_H37Rv VVGHAVLHHIPDVELSLREVVRVLKPGGRFVFAG----------------
MUL_1900|M.ulcerans_Agy99 VVGHAVLHHIPDVELSLREVMRVLKPGGRFVFAG----------------
MSMEG_2329|M.smegmatis_MC2_155 VVGHAVLHHIPDVELSLREVVRILKPGGRFVFAG----------------
TH_2189|M.thermoresistible__bu VVGHAVLHHIPDVELSLREVVRVLRPGGRFVFAG----------------
MAV_3903|M.avium_104 VVGHAVLHHIPDVELSLREVIRVLRPGGRFVFAG----------------
MLBr_01723|M.leprae_Br4923 VVGHAVLHHIPDVELSLREVLRVLKPGGRFVFAG----------------
MAB_3381c|M.abscessus_ATCC_199 VVGHAVLHHIPDVELSLREVLRVLKPGGRFVFAG----------------
Mvan_3244|M.vanbaalenii_PYR-1 VFDFGIIHHIPDWRAAVAEAARVLIPGGRFYFE-----------------
Mflv_2862|M.gilvum_PYR-GCK VFDFGIVHHIPDWRNAVAEVARVLAPGGRFYFD-----------------
MMAR_0694|M.marinum_M VFSQAVLYHVHDRERALAEVARVLQPQGLFVFDDLVTPQRPVSALAREVV
*.. .:::*: * . :: *. *:* * * * *
Mb3064c|M.bovis_AF2122/97 ------EPTTVGDGYARTLSTLTWRVVTNATKLPGLRGWRRPQGELDESS
Rv3038c|M.tuberculosis_H37Rv ------EPTTVGDGYARTLSTLTWRVVTNATKLPGLRGWRRPQGELDESS
MUL_1900|M.ulcerans_Agy99 ------EPTTVGNEYARTLSTLTWRIATNVTKLPGLGGWRRPQEELDESS
MSMEG_2329|M.smegmatis_MC2_155 ------EPTTVGNRYARELSTLTWHATTNLTKLPFLSGWRRPQEELDESS
TH_2189|M.thermoresistible__bu ------EPTTVGNSYARALSTLTWHVATTVTRLPGLQGWRRPQAELDESS
MAV_3903|M.avium_104 ------EPTSAGDVYARELSTLTWRIATNVTKLPGLGSWRRPQAELDESS
MLBr_01723|M.leprae_Br4923 ------EPTDVGNRYARVFADLTWKVTIRVVQLPGLSAWRRTQVEIDENS
MAB_3381c|M.abscessus_ATCC_199 ------EPTTVGNLYARALADLTWKATVQAMKLPGLESWRRPQEELDENS
Mvan_3244|M.vanbaalenii_PYR-1 ------------------------EVTAHALSRP---TYRLLFDHPTDDR
Mflv_2862|M.gilvum_PYR-GCK ------------------------EVTAHALARP---SYKRLFDHPTEDR
MMAR_0694|M.marinum_M YDRLLFEPTFSADDYRKALARHGFVVYETRDLSPHLAQSYKLLADLAEPS
* . :
Mb3064c|M.bovis_AF2122/97 RAAALEALVDL--------------------------------------H
Rv3038c|M.tuberculosis_H37Rv RAAALEALVDL--------------------------------------H
MUL_1900|M.ulcerans_Agy99 RAAALEAIVDL--------------------------------------H
MSMEG_2329|M.smegmatis_MC2_155 RAAALEAVVDL--------------------------------------H
TH_2189|M.thermoresistible__bu RAAALEAVVDL--------------------------------------H
MAV_3903|M.avium_104 RAAALEAIVDL--------------------------------------H
MLBr_01723|M.leprae_Br4923 RAAALEWVVDL--------------------------------------H
MAB_3381c|M.abscessus_ATCC_199 RAAALEWVVDL--------------------------------------H
Mvan_3244|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2862|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0694|M.marinum_M CPRLATAYRQIPEVIQDAEVGWSFFLGKKVVDRLEWIYNSDPDLSLEQKY
Mb3064c|M.bovis_AF2122/97 TFTPQDLQRIAHNAGAVEVQTATEEFTAAMLGWPLRTFECTVPPGRLGWG
Rv3038c|M.tuberculosis_H37Rv TFTPQDLQRIAHNAGAVEVQTATEEFTAAMLGWPLRTFECTVPPGRLGWG
MUL_1900|M.ulcerans_Agy99 TFAPEDLERMAKNAGAIRVETASEEFSAAMFGWPVRTFESSVPPGRLGWN
MSMEG_2329|M.smegmatis_MC2_155 TFDPADLERMATNAGAVEVRTASEEFTAAMLGWPVRTFEAAVPPGRLGWG
TH_2189|M.thermoresistible__bu TFEPSDLERMARNAGAVEVATASEEFTAAMLGWPVRTFEAAVPPGKLGWN
MAV_3903|M.avium_104 TFTPGDLERMAANAGATEVRTVSEEFTAAMFGWPVRTFEASVPPGRLGWG
MLBr_01723|M.leprae_Br4923 TFEPKVLETMATNAGAVQVKTVTEEFTAAMLGWPLRTFESTMPPSKLGWG
MAB_3381c|M.abscessus_ATCC_199 TFDPADLEKMAANAGAVAVRTASEEFTAAMLGWPVRTFESTVPPGKLGWG
Mvan_3244|M.vanbaalenii_PYR-1 -FTAEDFLDELGRHGLVILGSVTRIQGDYLLGVAAKPLAADGGR------
Mflv_2862|M.gilvum_PYR-GCK -FSAEQFSDELAHHGLVVLGSRTRIQGDYLLGVAAKPLPTGCE-------
MMAR_0694|M.marinum_M DAWAESYDSDLRDNYQENPRRAAALLAAHLPERSAPVLDAGAGTGLVGEE
. : : . :
Mb3064c|M.bovis_AF2122/97 WARFAFTSWKTLGWVDANVWRHVVPKGWFYNVMITGVKPS----------
Rv3038c|M.tuberculosis_H37Rv WARFAFTSWKTLGWVDANVWRHVVPKGWFYNVMITGVKPS----------
MUL_1900|M.ulcerans_Agy99 WARFAFNGWKALGWMDANVWRHVAPKGWFYNVMVTGVKPS----------
MSMEG_2329|M.smegmatis_MC2_155 WAKFAFNSWTTLSWVDANVWRKVVPKGWFYNVMVTGVKPS----------
TH_2189|M.thermoresistible__bu WAKFAFGSWTALSWVDANVWRRVMPKGWFYNVMVTGVKPS----------
MAV_3903|M.avium_104 WAKFAFNGWKTLSWVDANIWRRVVPKGWFYNVMVTGVKPT----------
MLBr_01723|M.leprae_Br4923 WARFAFTSWKTLSWVDANVWRRVIPKSWFYNIIITGVKPS----------
MAB_3381c|M.abscessus_ATCC_199 WAKFAFNGWKALSWVDENVWRHVVPKGWYYNVMITGIKPS----------
Mvan_3244|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2862|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0694|M.marinum_M LHKLGYDNLTALDRSLG-MLSHARAKNVYRDVVHGALAAAPTLFPDQRFA
Mb3064c|M.bovis_AF2122/97 --------------------------------------------------
Rv3038c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1900|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2329|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu --------------------------------------------------
MAV_3903|M.avium_104 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
MAB_3381c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3244|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2862|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0694|M.marinum_M AIVAVGVFTFAHAGPEDLAALDRVLEPGGWLVIAVRADYLDQHRTLEQTL
Mb3064c|M.bovis_AF2122/97 -------------------------------------
Rv3038c|M.tuberculosis_H37Rv -------------------------------------
MUL_1900|M.ulcerans_Agy99 -------------------------------------
MSMEG_2329|M.smegmatis_MC2_155 -------------------------------------
TH_2189|M.thermoresistible__bu -------------------------------------
MAV_3903|M.avium_104 -------------------------------------
MLBr_01723|M.leprae_Br4923 -------------------------------------
MAB_3381c|M.abscessus_ATCC_199 -------------------------------------
Mvan_3244|M.vanbaalenii_PYR-1 -------------------------------------
Mflv_2862|M.gilvum_PYR-GCK -------------------------------------
MMAR_0694|M.marinum_M AKLGYRIEARDEYEIFDAEPMVALGCRKPINDDGASR