For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ELSGKTVLVTGATGGLGRAIASSLADRGAELILSSRKPAELDQLAASLPGRNHRSVVSDLAEPGAALALL AEAGEIDVLVANAALPASGKLETFTAEQVDRALRVNLEIPVQMTRELIPAFTQRGSGHFVFVSSISGKTA TARASLYAATKFGIRGFALCLRDDLRPAGVGVSVVSPGAISGAGMLADSGATAPPLIGTGRPDQVGAAVV TAIERNRGEVTVAPLRQRALARFAANAPEIASRLGGGIAAKAADEIAAGQLHKR*
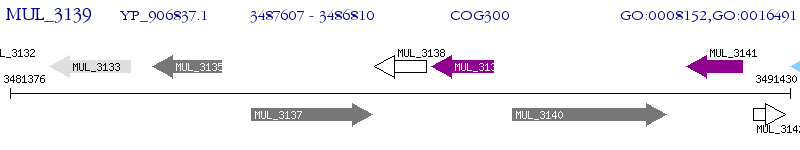
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3139 | - | - | 100% (265) | short-chain dehydrogenase |
| M. ulcerans Agy99 | MUL_0819 | adh_1 | 4e-26 | 34.07% (226) | short-chain dehydrogenase Adh_1 |
| M. ulcerans Agy99 | MUL_3400 | - | 5e-22 | 37.77% (188) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1079 | - | 3e-23 | 31.08% (251) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_0296 | - | 1e-20 | 32.67% (251) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1050 | - | 3e-23 | 31.08% (251) | oxidoreductase |
| M. leprae Br4923 | MLBr_01740 | - | 2e-20 | 36.36% (187) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1526 | - | 2e-25 | 32.16% (227) | short chain dehydrogenase |
| M. marinum M | MMAR_2623 | - | 1e-141 | 98.49% (265) | short-chain dehydrogenase |
| M. avium 104 | MAV_4982 | - | 6e-22 | 28.11% (249) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_5204 | - | 5e-24 | 37.67% (223) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_2445 | - | 9e-24 | 31.64% (256) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4623 | - | 4e-26 | 32.92% (243) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1079|M.bovis_AF2122/97 ---MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAARREGALRR
Rv1050|M.tuberculosis_H37Rv ---MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAARREGALRR
MAB_1526|M.abscessus_ATCC_1997 MSRYPKIELAGAVVVITGGARGIGAATARLFADRGADVWIGDVDDVVAKE
TH_2445|M.thermoresistible__bu ------MTFAGKHALVTGAGSGIGAALCRALAAAGAEVMCTDIDGAAAER
MUL_3139|M.ulcerans_Agy99 ------MELSGKTVLVTGATGGLGRAIASSLADRGAELILSSRKPAELDQ
MMAR_2623|M.marinum_M ------MELSGKTVLVTGATGGLGRAIASSLADRGAELILSSRKPAELDQ
Mflv_0296|M.gilvum_PYR-GCK ----MDMGLSGKRALVTGSSSGLGRAIAESLAAEGASVVVHGRDEVRTGA
MSMEG_5204|M.smegmatis_MC2_155 -MNNHSTRLEGRTALVTGATGGLGVAIASSLAANGAHVVITGRDAARGDA
Mvan_4623|M.vanbaalenii_PYR-1 ------MELRGKRVLITGASRGIGESLAHNFAGAGARVALVARSRDSLES
MLBr_01740|M.leprae_Br4923 -MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRHCDGLAQ
MAV_4982|M.avium_104 ---------MPKTWFITGSSRGLGRALTTAVLDHGDHVVATARRPAQLDE
.:**. *:* : . * :
Mb1079|M.bovis_AF2122/97 VAREIEAA-GGRAMVAPLDVSSSES--VRAMVADVVGEFGRIDVVFNNAG
Rv1050|M.tuberculosis_H37Rv VAREIEAA-GGRAMVAPLDVSSSES--VRAMVADVVGEFGRIDVVFNNAG
MAB_1526|M.abscessus_ATCC_1997 TANGIPN-----AHSGALDVTKPES--WAGFLDQVRAESGPVDILINNAG
TH_2445|M.thermoresistible__bu TAQSIG------GRWAALDVTDAAA--VQAAVDGVVDRAGRLDLLFNNAG
MUL_3139|M.ulcerans_Agy99 LAASLP---GRNHRSVVSDLAEPG------AALALLAEAGEIDVLVANAA
MMAR_2623|M.marinum_M LAASLP---GRNHRSIVSDLAEPG------AALALLAEAGEIDVLVANAA
Mflv_0296|M.gilvum_PYR-GCK VADGIRAR-GGDAWAAVGDLATEG------GAGAVAAAAGEVDILVNNAG
MSMEG_5204|M.smegmatis_MC2_155 VVAGIRSA-GGRVDLVAADLAAGEDEVRRLARRAVEVAGGHIDILVNNAG
Mvan_4623|M.vanbaalenii_PYR-1 LAVELG------GTVHPADLSDSTQ--VSGLIRRVEDDGGPIDVLVNNAG
MLBr_01740|M.leprae_Br4923 TVSEARALGAQVPEYRALDISDYDE--VAAFGADIHARHPSMDIVMNIAG
MAV_4982|M.avium_104 LVDRYGPR----IRACELDVTDRAA--ARRAIAAAVTAFGRIDVVVNNAG
. *:: :*::. *.
Mb1079|M.bovis_AF2122/97 VSLVGPVDAETFLDDTREMLEIDYLGTVRVVREVLPIMKQQRSG-RIMNM
Rv1050|M.tuberculosis_H37Rv VSLVGPVDAETFLDDTREMLEIDYLGTVRVVREVLPIMKQQRSG-RIMNM
MAB_1526|M.abscessus_ATCC_1997 VMPLGGFIEEN-ERTTDLILDVNVRGPLNGARAVLPQMVARGRG-HVVNV
TH_2445|M.thermoresistible__bu IVWGGDTELLT-LDQWNAIIDVNIRGVVHGVAAAYPLMVRQGHG-HIVNT
MUL_3139|M.ulcerans_Agy99 LPAS-GKLETFTAEQVDRALRVNLEIPVQMTRELIPAFTQRGSG-HFVFV
MMAR_2623|M.marinum_M LPAS-GKLETFTAEQVDRALRVNLEIPVQMTRELIPAFTQRGSG-HFVFV
Mflv_0296|M.gilvum_PYR-GCK VYDG-LAWSDLAPRQWQSIYEVNVVSGVRMIEYLVPGMRQRGWG-RIIQI
MSMEG_5204|M.smegmatis_MC2_155 LWGMPEATSAVSEPDLLASFRTNVIAPFLLTGALAPAMAERGHG-AVVNV
Mvan_4623|M.vanbaalenii_PYR-1 IEST-ACFTDAPEDELRRVTDVNYLAPAELCRQAIPGMLRRGGG-HIVNV
MLBr_01740|M.leprae_Br4923 VSVW-GTVDRLTHDQWSNVVAINLMGPIHIIETFVPAILAAGRGGHLVNV
MAV_4982|M.avium_104 YANS-APVEEVSDDDFRAQVETNFFGVVNVTKAALPVLHRQRAG-HIVQI
: * : * .:
Mb1079|M.bovis_AF2122/97 SSVVGRKA-FARFAGYSSAMHAIAGFSDALRQELRGSGIAVSVIHPALTQ
Rv1050|M.tuberculosis_H37Rv SSVVGRKA-FARFAGYSSAMHAIAGFSDALRQELRGSGIAVSVIHPALTQ
MAB_1526|M.abscessus_ATCC_1997 ASMAGKLA-VPGMVTYNASKFGAVGLSVALRKEYGNTGVSVSCVLPSAVR
TH_2445|M.thermoresistible__bu ASMAGLAA-AGQLTSYVMTKHAVVGLSLALRSEAAAHGVGVLAVCPAAVE
MUL_3139|M.ulcerans_Agy99 SSISGKTA-TARASLYAATKFGIRGFALCLRDDLRPAGVGVSVVSPGAIS
MMAR_2623|M.marinum_M SSISGKTA-TARASLYAATKFGIRGFALCLRDDLRPAGVGVSVVSPGAIS
Mflv_0296|M.gilvum_PYR-GCK GGGLALQP-AADQPHYNATLAARHNLTVSLARELAGTGVTSNTVAPGAIL
MSMEG_5204|M.smegmatis_MC2_155 GSITGFIG-GDRAALYSSTKAAVHSLTKSWAVEYGPQGVRVNAVAPGPIA
Mvan_4623|M.vanbaalenii_PYR-1 SSLSGIVL-FPGLVSYSASKAALSHFTAGLRTDFRGLPIGTTLVELGPIP
MLBr_01740|M.leprae_Br4923 SSAAGLVA-LPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVVPGAVR
MAV_4982|M.avium_104 SSVGGRVGGSPGIAAYQASKFAVAGFSEALAAEVAPLGIKVTIAEPGALR
.. . * : . :: : : .
Mb1079|M.bovis_AF2122/97 TPLLANVDPADMP---------PPFRSLTPIPVHWVAAAVLDGVARRRAR
Rv1050|M.tuberculosis_H37Rv TPLLANVDPADMP---------PPFRSLTPIPVHWVAAAVLDGVARRRAR
MAB_1526|M.abscessus_ATCC_1997 TELASGAPLG------------KG---MPTVDPEDVAAAIVRSVDTRRAQ
TH_2445|M.thermoresistible__bu TPLLDKGAVGGFLGRD---YFIRGQGVRSAYDPDRLASDTLRAVQRNKAL
MUL_3139|M.ulcerans_Agy99 GAGMLADSGATAP------------PLIGTGRPDQVGAAVVTAIERNRGE
MMAR_2623|M.marinum_M GAGMFADSGATAP------------PLIGTGTPDQVGAAVVTAIERNRGE
Mflv_0296|M.gilvum_PYR-GCK TEPVREMTYRVAA------------QRGWGPTWEDVEKAAATTWFPNDIG
MSMEG_5204|M.smegmatis_MC2_155 TERVLEAADHVSG------------VLARIPS-----------------R
Mvan_4623|M.vanbaalenii_PYR-1 TDLLAKIDNYEPT-------------AKSFQRAYRWGVAVDTPRERVAEE
MLBr_01740|M.leprae_Br4923 TPLVDTIEIAGVDRQDPRFSRWVDRFRGHAVSAERAAEKILAGVAKNRYL
MAV_4982|M.avium_104 TEWAGSSMAVATVSPDYESTVGAMNRRRAGFNRNAPGDPARAAHAIIEVV
Mb1079|M.bovis_AF2122/97 VVVPFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYRGSVYRHQPT
Rv1050|M.tuberculosis_H37Rv VVVPFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYRGSVYRHQPT
MAB_1526|M.abscessus_ATCC_1997 ISV---PGWLAFGWGLVDLFVPEPVERLA----RRLIDDRRALTSLDLNA
TH_2445|M.thermoresistible__bu LVK---PRVAHAGW-LFARLAPGLMQRAS----IRFVERQRAG------Q
MUL_3139|M.ulcerans_Agy99 VTVA--PLRQRALARFAANAPEIASRLGG----------------GIAAK
MMAR_2623|M.marinum_M VTVA--PLRQRALARFAANAPEIASRLAG----------------GIAAK
Mflv_0296|M.gilvum_PYR-GCK RFGR--PEEVAAAVAFLAG--VHAGYISG----------------ADLRV
MSMEG_5204|M.smegmatis_MC2_155 RMST--PEEVAAAVVFLAG--DEAANIHG----------------AILSV
Mvan_4623|M.vanbaalenii_PYR-1 VVKAVLKGRRHVRLPKRSAPLAMLAEAPR--------------RIAEIIL
MLBr_01740|M.leprae_Br4923 IYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVFTRALRPSPATMRCVEPIK
MAV_4982|M.avium_104 IADDPPLRLLLGSDALSAALAKSHQQIAEAN------------RWAELSK
Mb1079|M.bovis_AF2122/97 ESAKAQAAQPERGYSSAR
Rv1050|M.tuberculosis_H37Rv ESAKAQAAQPERGYSSAR
MAB_1526|M.abscessus_ATCC_1997 RGAYIERIERQSKEHQK-
TH_2445|M.thermoresistible__bu RAAQVGR-----------
MUL_3139|M.ulcerans_Agy99 AADEIAAGQLHKR-----
MMAR_2623|M.marinum_M AADEIAAGQLHKR-----
Mflv_0296|M.gilvum_PYR-GCK DGGAIRSVA---------
MSMEG_5204|M.smegmatis_MC2_155 DGGWAAA-----------
Mvan_4623|M.vanbaalenii_PYR-1 TGVPHQAKS---------
MLBr_01740|M.leprae_Br4923 VGVYSQPRSGEGGKSGN-
MAV_4982|M.avium_104 SIDYPPVATPHRAARSR-