For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NNHSTRLEGRTALVTGATGGLGVAIASSLAANGAHVVITGRDAARGDAVVAGIRSAGGRVDLVAADLAAG EDEVRRLARRAVEVAGGHIDILVNNAGLWGMPEATSAVSEPDLLASFRTNVIAPFLLTGALAPAMAERGH GAVVNVGSITGFIGGDRAALYSSTKAAVHSLTKSWAVEYGPQGVRVNAVAPGPIATERVLEAADHVSGVL ARIPSRRMSTPEEVAAAVVFLAGDEAANIHGAILSVDGGWAAA*
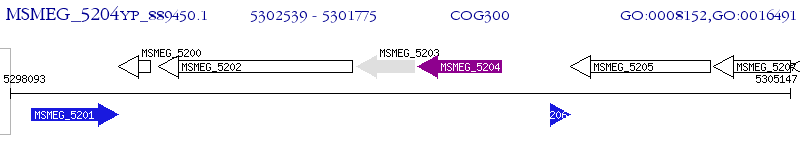
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5204 | - | - | 100% (254) | oxidoreductase, short chain dehydrogenase/reductase family protein |
| M. smegmatis MC2 155 | MSMEG_5383 | - | 7e-49 | 46.06% (241) | dehydrogenase/reductase SDR family protein member 4 |
| M. smegmatis MC2 155 | MSMEG_6753 | - | 5e-46 | 45.31% (245) | oxidoreductase, short chain dehydrogenase/reductase family |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0950c | - | 2e-34 | 40.73% (248) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0296 | - | 2e-35 | 40.23% (261) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0927c | - | 2e-34 | 40.73% (248) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 2e-22 | 30.99% (242) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4632 | - | 5e-31 | 34.40% (250) | short chain dehydrogenase |
| M. marinum M | MMAR_0629 | - | 6e-55 | 47.15% (246) | dehydrogenase |
| M. avium 104 | MAV_3012 | - | 2e-36 | 39.29% (252) | short-chain dehydrogenase/reductase SDR |
| M. thermoresistible (build 8) | TH_2488 | - | 8e-43 | 44.13% (247) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4442 | - | 5e-34 | 39.52% (248) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4593 | - | 3e-87 | 65.35% (254) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0950c|M.bovis_AF2122/97 ---------MILDMFR--LDDKVAVITGGGRGLGAAIALAFAQAGADVLI
Rv0927c|M.tuberculosis_H37Rv ---------MILDMFR--LDDKVAVITGGGRGLGAAIALAFAQAGADVLI
MUL_4442|M.ulcerans_Agy99 ---------MILDRFR--LDDQVAVITGGGRGFGAAIALAFAEAGADVVI
MAB_4632|M.abscessus_ATCC_1997 ---------MILDRFK--LDGNVAVVTGAGRGLGAAIAEAFAQAGADVLI
MAV_3012|M.avium_104 --------MTQHKHFDPLLSDRVAVVTGGGGGIGAATARLFAEHGAHVVI
MSMEG_5204|M.smegmatis_MC2_155 -----------MNNHSTRLEGRTALVTGATGGLGVAIASSLAANGAHVVI
Mvan_4593|M.vanbaalenii_PYR-1 -----------MSPNSTRLTGRTALVTGSTGGLGVTIAHALAADGAVVVV
MMAR_0629|M.marinum_M ------------------MAGLTALITGATSGIGRATAHGLAELGATVLV
TH_2488|M.thermoresistible__bu ----------------MEFQGLHVLVTGGTGGIGLACARAFAEAGAEVTV
Mflv_0296|M.gilvum_PYR-GCK --------------MDMGLSGKRALVTGSSSGLGRAIAESLAAEGASVVV
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
: . ::**. *:* : * :. * * :
Mb0950c|M.bovis_AF2122/97 ASRTSSELDAVAEQIRAAGRRAHTVAADLA--HPEVTAQLAGQAVGAFG-
Rv0927c|M.tuberculosis_H37Rv ASRTSSELDAVAEQIRAAGRRAHTVAADLA--HPEVTAQLAGQAVGAFG-
MUL_4442|M.ulcerans_Agy99 ASRTQSQLDEVADTVRSIGRRAHVVAADLA--HPEATAELAGQAVEAFG-
MAB_4632|M.abscessus_ATCC_1997 ASRTESQLREVAAKVQAAGRQAEVVVADLS--DTEASAALAQKAVDRFG-
MAV_3012|M.avium_104 ADIDAELADETASTIAASGGSAHPITADVR--NAAEVTHFARTVLDRYG-
MSMEG_5204|M.smegmatis_MC2_155 TGRDAARGDAVVAGIRSAGGRVDLVAADLA-AGEDEVRRLARRAVEVAGG
Mvan_4593|M.vanbaalenii_PYR-1 TGRDVARGESVVAGIRADGASAHFVAADLGGADEQAIDGLAQRAAEAASA
MMAR_0629|M.marinum_M SGRDEARGREVVDEVTARGGRGIFLAAELR--DVTGVRNLATAAADAGAG
TH_2488|M.thermoresistible__bu TGRDAGRG----AAAAGAHPRLRFIRGDLA--DLASVQNLIR-YADA---
Mflv_0296|M.gilvum_PYR-GCK HGRDEVRTGAVADGIRARGGDAWAAVGDLA-------TEGGAGAVAAAAG
MLBr_01807|M.leprae_Br4923 THRASVVPDWFFG-------VECDVTDNDA-------IGAAFKAVEEHQG
:
Mb0950c|M.bovis_AF2122/97 KLDIVVNNVGGTMPNT--LLSTSTKDLADAFAFNVGTAHALTVAAVPLML
Rv0927c|M.tuberculosis_H37Rv KLDIVVNNVGGTMPNT--LLSTSTKDLADAFAFNVGTAHALTVAAVPLML
MUL_4442|M.ulcerans_Agy99 RLDIVVNNVGGTMPNT--LLTTSTKDLKDAFTFNVATAHALTVAAVPLML
MAB_4632|M.abscessus_ATCC_1997 RLDVVVNNVGGTMPKP--FLDTTNDDLAEAFSFNVLSGHALLRAAVPHLL
MAV_3012|M.avium_104 RIDVLVNNVGHWLRHPGGFADTDPQLWDELYRINLHHVFLVTHAFLPTMI
MSMEG_5204|M.smegmatis_MC2_155 HIDILVNNAG-LWGMPEATSAVSEPDLLASFRTNVIAPFLLTGALAPAMA
Mvan_4593|M.vanbaalenii_PYR-1 GIDILVNNAA-KLLLPEPTATLSAGEVAEAFTVNVVAPMLLTGRLASAMA
MMAR_0629|M.marinum_M KVDILINSAG-AFPF-GPTADTSPDDFDAVFALNVRAPYFLVGALAPTMA
TH_2488|M.thermoresistible__bu -VDVLVNNAA-SFPG-ALTVEQDAESFTRTFATNVTGTYFLTAGLIPGML
Mflv_0296|M.gilvum_PYR-GCK EVDILVNNAG--VYDGLAWSDLAPRQWQSIYEVNVVSGVRMIEYLVPGMR
MLBr_01807|M.leprae_Br4923 PVEVLVANAG--ITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMI
::::: ... *: : :
Mb0950c|M.bovis_AF2122/97 EHSGGGSVINISSTMGRLAARGFAAYGTAKAALAHYTRLAALDLCPR-VR
Rv0927c|M.tuberculosis_H37Rv EHSGGGSVINISSTMGRLAARGFAAYGTAKAALAHYTRLAALDLCPR-VR
MUL_4442|M.ulcerans_Agy99 EHSGGGGIINITSTMGRVAGRGFAAYGTAKVALAHYTRLAALDLCPR-IR
MAB_4632|M.abscessus_ATCC_1997 K-SDNASVINITSTMGRLPGRAFLGYCTAKGALAHYTRTAAMDLSPR-IR
MAV_3012|M.avium_104 D-RGAGAIVNVSSVEGLRGYPEDPVYAAFKAAVIGFTRSLAVQVGNHGVR
MSMEG_5204|M.smegmatis_MC2_155 E-RGHGAVVNVGSITGFIGGDRAALYSSTKAAVHSLTKSWAVEYGPQGVR
Mvan_4593|M.vanbaalenii_PYR-1 A-RGSGSVVNIGSITGLRGTAGSAVYDATKASVHSLTRSWAAEYGPSGVR
MMAR_0629|M.marinum_M K-RGRGSIVNVTTMVAEFGAAGTGLYGASKAAIALLTKSWAAEYGPSGVR
TH_2488|M.thermoresistible__bu A-RGRGSIVNITSMVATKAVPGASVYSSSKAAVESLTRSWAVEFAAHGIR
Mflv_0296|M.gilvum_PYR-GCK Q-RGWGRIIQIGGGLALQPAADQPHYNATLAARHNLTVSLARELAGTGVT
MLBr_01807|M.leprae_Br4923 K-QRFGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSIT
. : : . * : . : * : :
Mb0950c|M.bovis_AF2122/97 VNAIAPGSILTSALEVVAAN--------------DELRAPMEQATPLRRL
Rv0927c|M.tuberculosis_H37Rv VNAIAPGSILTSALEVVAAN--------------DELRAPMEQATPLRRL
MUL_4442|M.ulcerans_Agy99 VNAIAPGSILTSALEVVAAN--------------DELRAPMEQATPLRRL
MAB_4632|M.abscessus_ATCC_1997 VNGIAPGSILTSALEVVAGN--------------DEMRNTLEQNTPLHRL
MAV_3012|M.avium_104 VNAVAPDVTESLQVPYSQWLS-------------AEEQSQWPRWVPVGRM
MSMEG_5204|M.smegmatis_MC2_155 VNAVAPGPIATER----VLEA-------------ADHVSGVLARIPSRRM
Mvan_4593|M.vanbaalenii_PYR-1 VNAVAPGPIATER----AAEF-------------AEHIAPALARVPSRRM
MMAR_0629|M.marinum_M VNAVSPGPTRTEG----TVEM-------------GEALDQLAAAAPAGRP
TH_2488|M.thermoresistible__bu VNAVAPGPTSTDGV---ATEW-------------GETNEALGRALPIGRT
Mflv_0296|M.gilvum_PYR-GCK SNTVAPGAILTEPVREMTYRVAAQRGWGPTWEDVEKAAATTWFPNDIGRF
MLBr_01807|M.leprae_Br4923 ANVVAPGFIDTEMTRAMDS----------------KYQEAALQAIPLGRI
* ::*. : . *
Mb0950c|M.bovis_AF2122/97 GDPVDIAAAAVYLASPAGSFLTGKTLEVDGGLTFPN-----LDLPIPDL-
Rv0927c|M.tuberculosis_H37Rv GDPVDIAAAAVYLASPAGSFLTGKTLEVDGGLTFPN-----LDLPIPDL-
MUL_4442|M.ulcerans_Agy99 GDPVDIAAAAVYLASPAGSFLTGKTLEVDGGLTHPN-----LDIPVPDL-
MAB_4632|M.abscessus_ATCC_1997 GDPADIAAAAVYLASPAGSFLTGKVLEVDGGLIAPN-----LDIPLPDLA
MAV_3012|M.avium_104 GLPEDQARVILFLASDCSSFITGHTIPTDGGTTAAGGWFRSSRRPGREWT
MSMEG_5204|M.smegmatis_MC2_155 STPEEVAAAVVFLAGDEAANIHGAILSVDGGWAAA---------------
Mvan_4593|M.vanbaalenii_PYR-1 STPQEVAAAVVFLASDDAANIQGAVLSVDGGWAAT---------------
MMAR_0629|M.marinum_M ADPTEIASTIVYLASDAASFIHGAVVPVDGGRAAV---------------
TH_2488|M.thermoresistible__bu ADPDEIAQAVVFLASSRASFITGAVLHVDGGGIAV---------------
Mflv_0296|M.gilvum_PYR-GCK GRPEEVAAAVAFLAGVHAGYISGADLRVDGGAIRSVA-------------
MLBr_01807|M.leprae_Br4923 GAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH--------------
. : * . :**. .. : * : .***
Mb0950c|M.bovis_AF2122/97 ------
Rv0927c|M.tuberculosis_H37Rv ------
MUL_4442|M.ulcerans_Agy99 ------
MAB_4632|M.abscessus_ATCC_1997 ------
MAV_3012|M.avium_104 NRPADP
MSMEG_5204|M.smegmatis_MC2_155 ------
Mvan_4593|M.vanbaalenii_PYR-1 ------
MMAR_0629|M.marinum_M ------
TH_2488|M.thermoresistible__bu ------
Mflv_0296|M.gilvum_PYR-GCK ------
MLBr_01807|M.leprae_Br4923 ------