For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSRYPKIELAGAVVVITGGARGIGAATARLFADRGADVWIGDVDDVVAKETANGIPNAHSGALDVTKPES WAGFLDQVRAESGPVDILINNAGVMPLGGFIEENERTTDLILDVNVRGPLNGARAVLPQMVARGRGHVVN VASMAGKLAVPGMVTYNASKFGAVGLSVALRKEYGNTGVSVSCVLPSAVRTELASGAPLGKGMPTVDPED VAAAIVRSVDTRRAQISVPGWLAFGWGLVDLFVPEPVERLARRLIDDRRALTSLDLNARGAYIERIERQS KEHQK
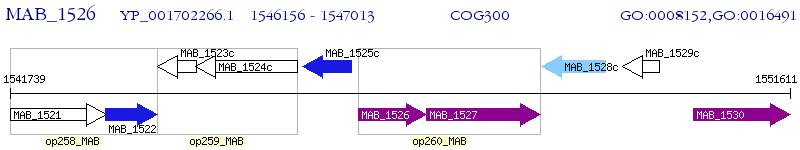
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1526 | - | - | 100% (285) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3405 | - | 7e-55 | 44.65% (271) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4335 | - | 2e-49 | 41.48% (270) | putative short-chain dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1896c | - | 2e-51 | 42.05% (264) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0217 | - | 4e-48 | 40.22% (271) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv1865c | - | 3e-51 | 42.05% (264) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 4e-30 | 31.93% (238) | short chain alcohol dehydrogenase |
| M. marinum M | MMAR_2743 | - | 2e-52 | 42.24% (277) | short-chain type dehydrogenase |
| M. avium 104 | MAV_2854 | - | 2e-52 | 42.59% (270) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0771 | - | 5e-48 | 40.15% (274) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3427 | - | 7e-52 | 45.09% (275) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3011 | - | 7e-52 | 41.88% (277) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0690 | - | 5e-50 | 40.73% (275) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0217|M.gilvum_PYR-GCK MDN-------IR----GKTIAITGAARGIGYATAEALLAHGARVVIGDRD
MSMEG_0771|M.smegmatis_MC2_155 MDN-------IR----GKTIAITGAARGIGYATAKALLAHGARVVIGDRD
Mvan_0690|M.vanbaalenii_PYR-1 MDN-------IR----GKTIAITGAARGIGYATAEALLRRGARVVIGDRD
TH_3427|M.thermoresistible__bu MDN-------IR----GKTIAITGAARGIGYATATALLARGARVVIGDRD
Mb1896c|M.bovis_AF2122/97 MPGRTSIGVKIRDKVQDKVIAITGGARGIGLATAAALHNLGAKVAIGDID
Rv1865c|M.tuberculosis_H37Rv MPGRTSIGVKIRDKVQDKVIAITGGARGIGLATAAALHNLGAKVAIGDID
MMAR_2743|M.marinum_M MADSTTIGVRVR----DKVIVITGGARGIGLATATALHKLGAKVAIGDVD
MUL_3011|M.ulcerans_Agy99 MADSTTIGVRVR----NKVIVITGGARGIGLATATALHKLGAKVAIGDVG
MAV_2854|M.avium_104 MADTASIGLKVR----DKVVVITGGARGIGLATATALHKLGAKVAIGDID
MAB_1526|M.abscessus_ATCC_1997 MSR------YPKIELAGAVVVITGGARGIGAATARLFADRGADVWIGDVD
MLBr_01094|M.leprae_Br4923 MEG-----------FAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVD
* . . .:**.. *** * * : ** : *.* .
Mflv_0217|M.gilvum_PYR-GCK VALQESSVAKLTNLGPVSGY-PLDVTDQESFATFLDKARTDGGGHIDVLI
MSMEG_0771|M.smegmatis_MC2_155 VALQESAVVELSKLGQVSGY-PLDVTDRESFATFLDKARTDGGGHIDVLI
Mvan_0690|M.vanbaalenii_PYR-1 VALQESAVAQLTKLGPVSGY-PLDVTDRDSFATFLDKARTDGGGHVDVLI
TH_3427|M.thermoresistible__bu VALQESAVAQLGRLGPVSGY-PLDVTDRESFATFLDKARTDGGGHIDVLI
Mb1896c|M.bovis_AF2122/97 EAMAKESGADLD-LDMYG---KLDVTDPDSFSGFLDAVERQLG-PIDVLV
Rv1865c|M.tuberculosis_H37Rv EAMAKESGADLD-LDMYG---KLDVTDPDSFSGFLDAVERQLG-PIDVLV
MMAR_2743|M.marinum_M EPAVKEAGADLG-LEVYG---KLDVTDPNSFSDFLDQVERQLG-PLDVLV
MUL_3011|M.ulcerans_Agy99 EPAVKEAGADLG-LEVYG---KLDVTDPNSFSDFLDQVERQLG-PLDVLV
MAV_2854|M.avium_104 EVRVKESGAALD-LDVYG---KLDVTDPHSFSDFLDEVERQLG-PIDVLV
MAB_1526|M.abscessus_ATCC_1997 DVVAKETANGIP--NAHSG--ALDVTKPESWAGFLDQVRAESG-PVDILI
MLBr_01094|M.leprae_Br4923 GEGLAQTEGQLKAIGASARTDRLDVTEREAFLTYADVVHENFG-KVNQIY
.: : . ****. .:: : * .. : * :: :
Mflv_0217|M.gilvum_PYR-GCK NNAGVMPVGPFLDETEQSIRSSLEVNVYGVLTGCRLALPDMVKRRSGHII
MSMEG_0771|M.smegmatis_MC2_155 NNAGVMPVGPFLEQSEQSIRSNIEVNVYGVLTGCQLALPDMVKRRRGHII
Mvan_0690|M.vanbaalenii_PYR-1 NNAGVMPIGPFLDQSEQSIRSSIEVNLYGVITGCQLALPDMIARRRGHII
TH_3427|M.thermoresistible__bu NNAGVMPIGPFLEQSEQAIRSSIEVNLYGVLTGCQLALPDMVRRRSGHII
Mb1896c|M.bovis_AF2122/97 NNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQRMVPRGRGHVI
Rv1865c|M.tuberculosis_H37Rv NNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQRMVPRGRGHVI
MMAR_2743|M.marinum_M NNAGIMPVGRIVDEPDSVTRRILDINVYGVMVGSKLAAQRMVPRGRGHVI
MUL_3011|M.ulcerans_Agy99 NNAGIMPVGRIVDEPDSVTRRILDINVYGVMVGSKLAAQRMVPRGRGHVI
MAV_2854|M.avium_104 NNAGIMPLGRVVDESDAVTRRILDINVYGVILGSKLALARMIPRGRGHVI
MAB_1526|M.abscessus_ATCC_1997 NNAGVMPLGGFIEENERTTDLILDVNVRGPLNGARAVLPQMVARGRGHVV
MLBr_01094|M.leprae_Br4923 NNAGIAFTGDVEVSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVI
****: * . . :::: * : * : :: **::
Mflv_0217|M.gilvum_PYR-GCK NIASLSGVIPLPGQVVYVGAKYAVVGLTTALADEMAPHG--VNVSVIMPP
MSMEG_0771|M.smegmatis_MC2_155 NIASLSGVIPLPGQVVYVGAKYAVVGLSTALADEMAPYG--VDVSVIMPP
Mvan_0690|M.vanbaalenii_PYR-1 NIASLSGLIPVPGQVVYVGAKFGVVGLSTALADEVAPQG--VDVSVIMPP
TH_3427|M.thermoresistible__bu NIASMSGLIPVPGQVPYNAAKFGVVGLSVALADEMAPYG--VEVSVVMPP
Mb1896c|M.bovis_AF2122/97 NVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSAG--VKFSMVLPS
Rv1865c|M.tuberculosis_H37Rv NVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSAG--VKFSMVLPS
MMAR_2743|M.marinum_M NVASLAGEIYVVGLATYCASKHAVIAFTDAARIEYRSTG--VKFSMVLPT
MUL_3011|M.ulcerans_Agy99 NVASLAGEIYVVGLATYCASKHAVIAFTDAARIEYRTTG--VKFSMVLPT
MAV_2854|M.avium_104 NVASLAGETYLAGAATYCASKHAVVGFTDAARIEYRRSG--VKFSVVKPT
MAB_1526|M.abscessus_ATCC_1997 NVASMAGKLAVPGMVTYNASKFGAVGLSVALRKEYGNTG--VSVSCVLPS
MLBr_01094|M.leprae_Br4923 NISSVFGLFSVPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPG
*::*: * * . * .:*... .:: : * * *..: : *
Mflv_0217|M.gilvum_PYR-GCK FTNTELIAGTKSGGAIKPVEPEDIAAAIVKTLNKPKTHVSVPPPLRFTAQ
MSMEG_0771|M.smegmatis_MC2_155 FTNTDLISGTKSGGAIKPVEPEEIAAAIVKTLNKPKTHVSVPPPLRFTAQ
Mvan_0690|M.vanbaalenii_PYR-1 FTNTDLISGTTSSGALKPVEPEDIAAAVVKTLNKPKTHVSVPPPLRFTAQ
TH_3427|M.thermoresistible__bu FTRTELISGTKETAATKPAEPEDIAAAIVKTLDKPKTHVSVPGPLRFIAQ
Mb1896c|M.bovis_AF2122/97 FVNTELIAGTGGIKGFKNAEPADIADAIVGLIVHPKPRVRVTKAAGSMIV
Rv1865c|M.tuberculosis_H37Rv FVNTELIAGTGGIKGFKNAEPADIADAIVGLIVHPKPRVRVTKAAGSMIV
MMAR_2743|M.marinum_M FVNTELASGTPGMKGFKNAEPSDIADAIVALVANPKPRVRVTKAAGAMIA
MUL_3011|M.ulcerans_Agy99 FVNTELASGTPGMKGFKNAEPSDIADAIVALVANPKPRVRVTKAAGAMIA
MAV_2854|M.avium_104 FVNTELIAGTSGAKGVRNAEPSDIADAIVKLVAHPRPRVRVTRTAGAIIA
MAB_1526|M.abscessus_ATCC_1997 AVRTELASGAPLGKGMPTVDPEDVAAAIVRSVDTRRAQISVPGWLAFGWG
MLBr_01094|M.leprae_Br4923 GIKTAIARNATAAEGLDVSKIASRFDTWVAHTSPQHAARIILKAVRKKKA
.* : .: . . . : * :. :
Mflv_0217|M.gilvum_PYR-GCK AAQMLPPKGRRAMNKALGLDTVFLD-VDTDKRKGYEERARAARGVVEGSQ
MSMEG_0771|M.smegmatis_MC2_155 AAQMLPPKGRRWLNKKLGLDNVFLE-FDTAKRKAYEDRAQAALGVVESEK
Mvan_0690|M.vanbaalenii_PYR-1 AAQMLPPRGRRWLSRKLGLDRVFLE-FDTTKRQGYEQRAQSALGVVEGSE
TH_3427|M.thermoresistible__bu AAQTLGPRGRRWLNKRLGLDTVFLE-FDQQARQGYEQRAQQALGVLEGGN
Mb1896c|M.bovis_AF2122/97 AQRFMPRQVSEGLNRLLGGEHVFTDDVDMEKRRTYEARARGEE-------
Rv1865c|M.tuberculosis_H37Rv AQRFMPRQVSEGLNRLLGGEHVFTDDVDMEKRRTYEARARGEE-------
MMAR_2743|M.marinum_M SQKYLPRWLSEGLNRLLGGEHVFTDDVDAAKREAYEARARGETGES----
MUL_3011|M.ulcerans_Agy99 SQKYLPRWLSEGLNRLLGGEHVFTDDVDAAKREAYEARARGETGES----
MAV_2854|M.avium_104 SQKFMPRALSEGLNRLLGGEHVFTDAVDVEKRQAYEARARGEQ-------
MAB_1526|M.abscessus_ATCC_1997 LVDLFVPEPVERLARRLIDDRRALTSLDLNARGAYIERIERQSKEHQK--
MLBr_01094|M.leprae_Br4923 RVLVGPDAKVANVVVRFSGGAGYQRLFAQVASRLILNQR-----------
: : . :
Mflv_0217|M.gilvum_PYR-GCK G---------
MSMEG_0771|M.smegmatis_MC2_155 K---------
Mvan_0690|M.vanbaalenii_PYR-1 NKPSPKM---
TH_3427|M.thermoresistible__bu KQQDRSGQKS
Mb1896c|M.bovis_AF2122/97 ----------
Rv1865c|M.tuberculosis_H37Rv ----------
MMAR_2743|M.marinum_M ----------
MUL_3011|M.ulcerans_Agy99 ----------
MAV_2854|M.avium_104 ----------
MAB_1526|M.abscessus_ATCC_1997 ----------
MLBr_01094|M.leprae_Br4923 ----------