For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MELSGKTVLVTGATGGLGRAIASSLADRGAELILSSRKPAELDQLAASLPGRNHRSIVSDLAEPGAALAL LAEAGEIDVLVANAALPASGKLETFTAEQVDRALRVNLEIPVQMTRELIPAFTQRGSGHFVFVSSISGKT ATARASLYAATKFGIRGFALCLRDDLRPAGVGVSVVSPGAISGAGMFADSGATAPPLIGTGTPDQVGAAV VTAIERNRGEVTVAPLRQRALARFAANAPEIASRLAGGIAAKAADEIAAGQLHKR
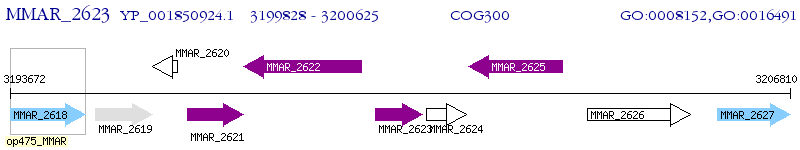
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2623 | - | - | 100% (265) | short-chain dehydrogenase |
| M. marinum M | MMAR_1636 | - | 6e-22 | 37.77% (188) | short chain alcohol dehydrogenase/reductase |
| M. marinum M | MMAR_4195 | - | 3e-21 | 29.92% (264) | short-chain type dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1079 | - | 1e-23 | 29.96% (257) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4313 | - | 3e-20 | 35.64% (188) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv1050 | - | 1e-23 | 29.96% (257) | oxidoreductase |
| M. leprae Br4923 | MLBr_01740 | - | 2e-20 | 36.36% (187) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1526 | - | 2e-25 | 32.16% (227) | short chain dehydrogenase |
| M. avium 104 | MAV_4982 | - | 6e-22 | 28.92% (249) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_5204 | - | 4e-24 | 37.22% (223) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_2445 | - | 2e-23 | 31.25% (256) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3139 | - | 1e-141 | 98.49% (265) | short-chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4623 | - | 5e-25 | 32.10% (243) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2623|M.marinum_M -------MELSGKTVLVTGATGGLGRAIASSLADRGAELILSS-------
MUL_3139|M.ulcerans_Agy99 -------MELSGKTVLVTGATGGLGRAIASSLADRGAELILSS-------
Mb1079|M.bovis_AF2122/97 ----MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAARREGALR
Rv1050|M.tuberculosis_H37Rv ----MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAARREGALR
Mflv_4313|M.gilvum_PYR-GCK MAQRSGSDRFSGKRCFVTGAASGIGRATALKLAARGAELYLTDRDADGLA
MLBr_01740|M.leprae_Br4923 MVKT--AQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRHCDGLA
MAB_1526|M.abscessus_ATCC_1997 -MSRYPKIELAGAVVVITGGARGIGAATARLFADRGADVWIGD-----VD
TH_2445|M.thermoresistible__bu -------MTFAGKHALVTGAGSGIGAALCRALAAAGAEVMCTD-----ID
Mvan_4623|M.vanbaalenii_PYR-1 -------MELRGKRVLITGASRGIGESLAHNFAGAGARVALVAR-----S
MAV_4982|M.avium_104 ----------MPKTWFITGSSRGLGRALTTAVLDHGDHVVATAR----RP
MSMEG_5204|M.smegmatis_MC2_155 --MNNHSTRLEGRTALVTGATGGLGVAIASSLAANGAHVVITGRDAARGD
.:**. *:* : . * :
MMAR_2623|M.marinum_M RKPAELDQLAASLPGRNHRSIVSDLAEPGAALALLAEAG-EIDVLVANAA
MUL_3139|M.ulcerans_Agy99 RKPAELDQLAASLPGRNHRSVVSDLAEPGAALALLAEAG-EIDVLVANAA
Mb1079|M.bovis_AF2122/97 RVAREIEAAGGR-AMVAPLDVSSSESVRAMVADVVGEFG-RIDVVFNNAG
Rv1050|M.tuberculosis_H37Rv RVAREIEAAGGR-AMVAPLDVSSSESVRAMVADVVGEFG-RIDVVFNNAG
Mflv_4313|M.gilvum_PYR-GCK LTVADARALGGVVAEHRALDISDHDAVVAFADDIHSRHP-AMDVVMNIAG
MLBr_01740|M.leprae_Br4923 QTVSEARALGAQVPEYRALDISDYDEVAAFGADIHARHP-SMDIVMNIAG
MAB_1526|M.abscessus_ATCC_1997 DVVAKETANGIPNAHSGALDVTKPESWAGFLDQVRAESG-PVDILINNAG
TH_2445|M.thermoresistible__bu GAAAERTAQSIG-GRWAALDVTDAAAVQAAVDGVVDRAG-RLDLLFNNAG
Mvan_4623|M.vanbaalenii_PYR-1 RDSLESLAVELG-GTVHPADLSDSTQVSGLIRRVEDDGG-PIDVLVNNAG
MAV_4982|M.avium_104 AQLDELVDRYGPRIRACELDVTDRAAARRAIAAAVTAFG-RIDVVVNNAG
MSMEG_5204|M.smegmatis_MC2_155 AVVAGIRSAGGRVDLVAADLAAGEDEVRRLARRAVEVAGGHIDILVNNAG
:*::. *.
MMAR_2623|M.marinum_M LPASGKLETFT-AEQVDRALRVNLEIPVQMTRELIPAFTQRGSG-HFVFV
MUL_3139|M.ulcerans_Agy99 LPASGKLETFT-AEQVDRALRVNLEIPVQMTRELIPAFTQRGSG-HFVFV
Mb1079|M.bovis_AF2122/97 VSLVGPVDAETFLDDTREMLEIDYLGTVRVVREVLPIMKQQRSG-RIMNM
Rv1050|M.tuberculosis_H37Rv VSLVGPVDAETFLDDTREMLEIDYLGTVRVVREVLPIMKQQRSG-RIMNM
Mflv_4313|M.gilvum_PYR-GCK ISAWGTVDKLT-HDDWTSLININLMGPIHVIETFVPPMMEARRGGHLVNV
MLBr_01740|M.leprae_Br4923 VSVWGTVDRLT-HDQWSNVVAINLMGPIHIIETFVPAILAAGRGGHLVNV
MAB_1526|M.abscessus_ATCC_1997 VMPLGGFIEEN-ERTTDLILDVNVRGPLNGARAVLPQMVARGRG-HVVNV
TH_2445|M.thermoresistible__bu IVWGGDTELLT-LDQWNAIIDVNIRGVVHGVAAAYPLMVRQGHG-HIVNT
Mvan_4623|M.vanbaalenii_PYR-1 IESTACFTDAP-EDELRRVTDVNYLAPAELCRQAIPGMLRRGGG-HIVNV
MAV_4982|M.avium_104 YANSAPVEEVS-DDDFRAQVETNFFGVVNVTKAALPVLHRQRAG-HIVQI
MSMEG_5204|M.smegmatis_MC2_155 LWGMPEATSAVSEPDLLASFRTNVIAPFLLTGALAPAMAERGHG-AVVNV
: * : * .:
MMAR_2623|M.marinum_M SSISG-KTATARASLYAATKFGIRGFALCLRDDLRPAGVGVSVVSPGAIS
MUL_3139|M.ulcerans_Agy99 SSISG-KTATARASLYAATKFGIRGFALCLRDDLRPAGVGVSVVSPGAIS
Mb1079|M.bovis_AF2122/97 SSVVG-RKAFARFAGYSSAMHAIAGFSDALRQELRGSGIAVSVIHPALTQ
Rv1050|M.tuberculosis_H37Rv SSVVG-RKAFARFAGYSSAMHAIAGFSDALRQELRGSGIAVSVIHPALTQ
Mflv_4313|M.gilvum_PYR-GCK SSAAG-IVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVVPGAVR
MLBr_01740|M.leprae_Br4923 SSAAG-LVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVVPGAVR
MAB_1526|M.abscessus_ATCC_1997 ASMAG-KLAVPGMVTYNASKFGAVGLSVALRKEYGNTGVSVSCVLPSAVR
TH_2445|M.thermoresistible__bu ASMAG-LAAAGQLTSYVMTKHAVVGLSLALRSEAAAHGVGVLAVCPAAVE
Mvan_4623|M.vanbaalenii_PYR-1 SSLSG-IVLFPGLVSYSASKAALSHFTAGLRTDFRGLPIGTTLVELGPIP
MAV_4982|M.avium_104 SSVGGRVGGSPGIAAYQASKFAVAGFSEALAAEVAPLGIKVTIAEPGALR
MSMEG_5204|M.smegmatis_MC2_155 GSITG-FIGGDRAALYSSTKAAVHSLTKSWAVEYGPQGVRVNAVAPGPIA
.* * * : . :: : : . .
MMAR_2623|M.marinum_M GAGMFADSGATAP------------PLIGTGTPDQVGAAVVTAIERNRGE
MUL_3139|M.ulcerans_Agy99 GAGMLADSGATAP------------PLIGTGRPDQVGAAVVTAIERNRGE
Mb1079|M.bovis_AF2122/97 TPLLANVDPADMP---------PPFRSLTPIPVHWVAAAVLDGVARRRAR
Rv1050|M.tuberculosis_H37Rv TPLLANVDPADMP---------PPFRSLTPIPVHWVAAAVLDGVARRRAR
Mflv_4313|M.gilvum_PYR-GCK TPLVDTVRIAGVDRDDPRVQKWTNRFSGHAVSPDVAADKILDGVARNRYL
MLBr_01740|M.leprae_Br4923 TPLVDTIEIAGVDRQDPRFSRWVDRFRGHAVSAERAAEKILAGVAKNRYL
MAB_1526|M.abscessus_ATCC_1997 TELASGAPLG------------KG---MPTVDPEDVAAAIVRSVDTRRAQ
TH_2445|M.thermoresistible__bu TPLLDKGAVGGFLGRD---YFIRGQGVRSAYDPDRLASDTLRAVQRNKAL
Mvan_4623|M.vanbaalenii_PYR-1 TDLLAKIDNYEPTAKS--FQRAYRWGVAVDTPRERVAEEVVKAVLKGRRH
MAV_4982|M.avium_104 TEWAGSSMAVATVSPDYESTVGAMNRRRAGFNRNAPGDPARAAHAIIEVV
MSMEG_5204|M.smegmatis_MC2_155 TERVLEAADHVSG--------------------------VLARIPSRRMS
.
MMAR_2623|M.marinum_M VTVAPLRQRALAR--FAANAPEIASRLAGGIAAKAADEIAAGQLHKR---
MUL_3139|M.ulcerans_Agy99 VTVAPLRQRALAR--FAANAPEIASRLGGGIAAKAADEIAAGQLHKR---
Mb1079|M.bovis_AF2122/97 VVVPFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYRGSVYRHQPT
Rv1050|M.tuberculosis_H37Rv VVVPFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYRGSVYRHQPT
Mflv_4313|M.gilvum_PYR-GCK IYTSLDIRALYL--FKRTMWLPYSIAMKQVNPLFTRALRPK---------
MLBr_01740|M.leprae_Br4923 IYTSHDIRVLYG--FKRLAWWPYGVVMKQVNVVFTRALRPSPATMRCVEP
MAB_1526|M.abscessus_ATCC_1997 ISVPGWLAFGWG--LVDLFVPEPVERLARRLIDDRRALTSLDLNARGAYI
TH_2445|M.thermoresistible__bu LVKPRVAHAGW---LFARLAPGLMQRASIRFVERQRAG------QRAAQV
Mvan_4623|M.vanbaalenii_PYR-1 VRLPKRS-------APLAMLAEAPRRIAEIILTGVPHQAKS---------
MAV_4982|M.avium_104 IADDPPLRLLLGSDALSAALAKSHQQIAEANRWAELSKSIDYPPVATPHR
MSMEG_5204|M.smegmatis_MC2_155 TPEEVAAAVVFLAGDEAANIHGAILSVDGGWAAA----------------
MMAR_2623|M.marinum_M -------------------
MUL_3139|M.ulcerans_Agy99 -------------------
Mb1079|M.bovis_AF2122/97 ESAKAQAAQPERGYSSAR-
Rv1050|M.tuberculosis_H37Rv ESAKAQAAQPERGYSSAR-
Mflv_4313|M.gilvum_PYR-GCK ------TKQR---------
MLBr_01740|M.leprae_Br4923 IKVGVYSQPRSGEGGKSGN
MAB_1526|M.abscessus_ATCC_1997 ERIERQSKEHQK-------
TH_2445|M.thermoresistible__bu GR-----------------
Mvan_4623|M.vanbaalenii_PYR-1 -------------------
MAV_4982|M.avium_104 AARSR--------------
MSMEG_5204|M.smegmatis_MC2_155 -------------------