For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ELRGKRVLITGASRGIGESLAHNFAGAGARVALVARSRDSLESLAVELGGTVHPADLSDSTQVSGLIRRV EDDGGPIDVLVNNAGIESTACFTDAPEDELRRVTDVNYLAPAELCRQAIPGMLRRGGGHIVNVSSLSGIV LFPGLVSYSASKAALSHFTAGLRTDFRGLPIGTTLVELGPIPTDLLAKIDNYEPTAKSFQRAYRWGVAVD TPRERVAEEVVKAVLKGRRHVRLPKRSAPLAMLAEAPRRIAEIILTGVPHQAKS*
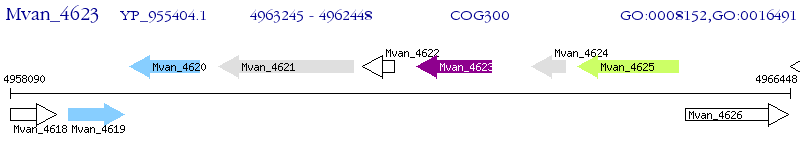
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4623 | - | - | 100% (265) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_0944 | - | 1e-28 | 38.12% (202) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2345 | - | 6e-27 | 34.17% (240) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1963c | - | 9e-27 | 39.11% (202) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0026 | - | 5e-30 | 41.09% (202) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv1928c | - | 9e-27 | 39.11% (202) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01740 | - | 4e-23 | 32.07% (237) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4335 | - | 2e-27 | 33.19% (238) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0254 | - | 4e-29 | 39.13% (184) | ketoacyl reductase |
| M. avium 104 | MAV_4597 | - | 3e-29 | 36.75% (234) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1073 | - | 1e-27 | 34.12% (255) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3427 | - | 3e-26 | 30.71% (254) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0819 | adh_1 | 7e-29 | 34.71% (242) | short-chain dehydrogenase Adh_1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAV_4597|M.avium_104 --------------------------MSKSPLRRFADQLVLATMRPPMAP
MSMEG_1073|M.smegmatis_MC2_155 --------------------------MSKHPLRRLTDNLFLASMRPPVTD
Mflv_0026|M.gilvum_PYR-GCK MRSSRKSASEVLLECEFGAGNRLRGVTSRNPLRRLADQVVLTSMRPPILP
Mvan_4623|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_0819|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0254|M.marinum_M --------------------------------------------------
MLBr_01740|M.leprae_Br4923 --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 -----MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQVAIAARHL
Rv1928c|M.tuberculosis_H37Rv -----MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQVAIAARHL
MAB_4335|M.abscessus_ATCC_1997 -------MGTAVQLAGKVVAVTGGARGIGRAIATAFAAEGAKVAIGDIDK
TH_3427|M.thermoresistible__bu ----------MDNIRGKTIAITGAARGIGYATATALLARGARVVIGDRDV
MAV_4597|M.avium_104 QVLVNRPLIKPVELAGKRVLLTGASSGIGEAAAEQFAREGARVVVVARRK
MSMEG_1073|M.smegmatis_MC2_155 QLIQLRAARDAVDLTGKRVLITGASSGIGAAAARKFAERGARVIVVARRA
Mflv_0026|M.gilvum_PYR-GCK ALLN-RPTRETIELRGKRVLLTGASSGIGEAAAEKLARRGATVVAVARRQ
Mvan_4623|M.vanbaalenii_PYR-1 -----------MELRGKRVLITGASRGIGESLAHNFAGAGARVALVARSR
MUL_0819|M.ulcerans_Agy99 ---------MSYTMAGRRVLITGASSGVGAALARMLAARGTTVGLIARRR
MMAR_0254|M.marinum_M -------MALPPPGKDRAAIVTGASSGIGEEFARILSQRGYQVVLVARSA
MLBr_01740|M.leprae_Br4923 ------MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRHC
.: :**.: *:* * * :
Mb1963c|M.bovis_AF2122/97 DALEKLADEIGTSGGKVVPVC-CDVSQHQQVTSMLDQVTAELG-GIDIAV
Rv1928c|M.tuberculosis_H37Rv DALEKLADEIGTSGGKVVPVC-CDVSQHQQVTSMLDQVTAELG-GIDIAV
MAB_4335|M.abscessus_ATCC_1997 KLCENTAAEIG---NSAIGLP-LDVTDHGSFEAFFDTIAATVG-PVDVIV
TH_3427|M.thermoresistible__bu ALQESAVAQLGRL-GPVSGYP-LDVTDRESFATFLDKARTDGGGHIDVLI
MAV_4597|M.avium_104 DLLDALAEGITRAGGEAIAMP-CDISDLDAADALVADVQQQLG-GVDILI
MSMEG_1073|M.smegmatis_MC2_155 ELLDEVAGGITASGGDAKARP-CDLTDLDAIDALVAEIESDHG-GLDILV
Mflv_0026|M.gilvum_PYR-GCK DLLDELVTRIADAGGDARAHA-CDLSDLDAVDELVATVEKDLG-GIDILV
Mvan_4623|M.vanbaalenii_PYR-1 DSLESLAVELG-----GTVHP-ADLSDSTQVSGLIRRVEDDGG-PIDVLV
MUL_0819|M.ulcerans_Agy99 DRLEEVLADCRKTSPQSVMWV-ADLCDTAGAARLALQAWQAMG-GIDVLV
MMAR_0254|M.marinum_M DRLEALAGRLG---SDTHPLP-ADLSVRSDRAGLVDRVA-ALGLVPDILI
MLBr_01740|M.leprae_Br4923 DGLAQTVSEARALGAQVPEYRALDISDYDEVAAFGADIHARHP-SMDIVM
*: : *: :
Mb1963c|M.bovis_AF2122/97 CNAGIITVTPMLD--MPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQGQGG
Rv1928c|M.tuberculosis_H37Rv CNAGIITVTPMLD--MPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQGQGG
MAB_4335|M.abscessus_ATCC_1997 NNAGIMPITPFDD--ESLESIQRQLDINVRGVMWGSQLAIKRMKPRG-GG
TH_3427|M.thermoresistible__bu NNAGVMPIGPFLE--QSEQAIRSSIEVNLYGVLTGCQLALPDMVRRR-SG
MAV_4597|M.avium_104 NNAGRSIRRPLAESLERWHDVERTMVLNYYAPLRLIRGIAPGMIERG-DG
MSMEG_1073|M.smegmatis_MC2_155 NNAGHSIRRPLADSLERWHDIERTMTLNYYSPLRLIRGFAPGMRERG-AG
Mflv_0026|M.gilvum_PYR-GCK NNAGRSIRRPLEESLERWHDVERTMTLNYYGPLRLIRGLAPGMLARG-DG
Mvan_4623|M.vanbaalenii_PYR-1 NNAGIESTACFTD--APEDELRRVTDVNYLAPAELCRQAIPGMLRRG-GG
MUL_0819|M.ulcerans_Agy99 NNAAIPKRREVSA--LRPTEVEQVMGVNFFAPMRMTLALLPRMLARG-SG
MMAR_0254|M.marinum_M NNAGLSTLGPVAK--SVPEQELNLVEVDVAAVVDLCSRFLPAMVERG-RG
MLBr_01740|M.leprae_Br4923 NIAGVSVWGTVDR--LTHDQWSNVVAINLMGPIHIIETFVPAILAAGRGG
*. . : . : *
Mb1963c|M.bovis_AF2122/97 VIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPHKIRVNSV
Rv1928c|M.tuberculosis_H37Rv VIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPHKIRVNSV
MAB_4335|M.abscessus_ATCC_1997 VIVNIASAAGKAG-FPGLAT-YCATKHAVVGLSESLSLEYEPAGISVVCV
TH_3427|M.thermoresistible__bu HIINIASMSGLIP-VPGQVP-YNAAKFGVVGLSVALADEMAPYGVEVSVV
MAV_4597|M.avium_104 HIINVSTWG-VLSEASPLFAVYNASKAALSTVSRVVETEWGDKGVHSTTL
MSMEG_1073|M.smegmatis_MC2_155 HIINVSTWG-VLSEASPLFSVYQASKAALSAVSRVIETEWGDGGVHSTTL
Mflv_0026|M.gilvum_PYR-GCK HIINVSTWG-VKTESPPLFGVYNASKAALSSVSRIIETEWSGRGVHATTL
Mvan_4623|M.vanbaalenii_PYR-1 HIVNVSSLS-GIVLFPGLVS-YSASKAALSHFTAGLRTDFRGLPIGTTLV
MUL_0819|M.ulcerans_Agy99 TIVNVSSVGGRLG--IIHESAYCASKFALCGWSESIAVDLAGSGVSVRLI
MMAR_0254|M.marinum_M AVLNVASVAGFAP--LPGQAAYGAAKAFVLSYTHSLRGELHGSGVSVTAL
MLBr_01740|M.leprae_Br4923 HLVNVSSAAGLVA--LPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVV
::* :: . * *:* : : : : : :
Mb1963c|M.bovis_AF2122/97 SPGYILTELVEPYTEYQPLW--EPKIP---------LGRLGRPEELAGLY
Rv1928c|M.tuberculosis_H37Rv SPGYILTELVEPYTEYQPLW--EPKIP---------LGRLGRPEELAGLY
MAB_4335|M.abscessus_ATCC_1997 MPGMVNTELISGLDTHWLLK--TVEPEDIANAVVKAVRRGVFPVFVPKMF
TH_3427|M.thermoresistible__bu MPPFTRTELISGTKETAATK--PAEPEDIAAAIVKTLDKPKTHVSVPGPL
MAV_4597|M.avium_104 YYPLVATPMIAPTKAYQGVP--ALTPEEAGR--WMITAARTRPVRIAPRM
MSMEG_1073|M.smegmatis_MC2_155 YYPLVDTPMIKPTRAFDGLP--ALSADEAAD--WMITAARHRPVQIAPRV
Mflv_0026|M.gilvum_PYR-GCK YYPLVKTPMIAPTRAYDGLP--GLSADEAAG--WIVTAARHRPVSIAPRI
Mvan_4623|M.vanbaalenii_PYR-1 ELGPIPTDLLAKIDNYEPTA--KSFQRAYRWGVAVDTPRERVAEEVVKAV
MUL_0819|M.ulcerans_Agy99 QPGPVDTEIWDRPDNEDPLYRGPKVPADLVAEGIIAAIGSDRFEHYLPEM
MMAR_0254|M.marinum_M SPGPVDTSFGDVAGFTQQET---ESLPKIMWKSVDQVAQAGIDGLAAGKA
MLBr_01740|M.leprae_Br4923 VPGAVRTPLVDTIEIAGVDRQDPRFSRWVDRFRGHAVSAERAAEKILAGV
* :
Mb1963c|M.bovis_AF2122/97 LYLASEASSYMTGSDIVIDGGYTCP-------------------------
Rv1928c|M.tuberculosis_H37Rv LYLASEASSYMTGSDIVIDGGYTCP-------------------------
MAB_4335|M.abscessus_ATCC_1997 GGMYRTMSVLPRSTGKFLTRKLGIQDFMLNAHGSDARKAYEDRASHSGPS
TH_3427|M.thermoresistible__bu RFIAQAAQTLGPRGRRWLNKRLGLDTVFLEFD-QQARQGYEQRAQQALGV
MAV_4597|M.avium_104 AIAAKALDTFGPRWVNAVMQRQTVQPNREAGA------------------
MSMEG_1073|M.smegmatis_MC2_155 AVGARALNAVAPGVVNAVMKRQSAQPDGVGRRPIFPAG------------
Mflv_0026|M.gilvum_PYR-GCK AVTAHAINSVAPGAVDTIMKRQRMQPRD----------------------
Mvan_4623|M.vanbaalenii_PYR-1 LKGRRHVRLPKRSAPLAMLAEAPRRIAEIILTGVPHQAKS----------
MUL_0819|M.ulcerans_Agy99 MALKDIVDAKNADLDTFLTAAAAMIWQLKALVYGVALEPFEIDEPANPLA
MMAR_0254|M.marinum_M VVVPGLANRISAELIRVTPPDWMGSIMARIHPAMKRQ-------------
MLBr_01740|M.leprae_Br4923 AKNRYLIYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVFTRALRPSPATMR
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 TE------------------------------------------------
TH_3427|M.thermoresistible__bu LEGGNKQQDRSGQKS-----------------------------------
MAV_4597|M.avium_104 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4623|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_0819|M.ulcerans_Agy99 CNLARTPTALRELADPRPLRPDWVITRPRLTGICGSDSKQILLDFGEQDA
MMAR_0254|M.marinum_M --------------------------------------------------
MLBr_01740|M.leprae_Br4923 CVEPIKVGVYSQPRSGEGGKSGN---------------------------
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAV_4597|M.avium_104 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4623|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_0819|M.ulcerans_Agy99 DNALAAFCSFPQVMGHEVVADVVELGPEAGGLEVGQRVVLNPWLSCRPRG
MMAR_0254|M.marinum_M --------------------------------------------------
MLBr_01740|M.leprae_Br4923 --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAV_4597|M.avium_104 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4623|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_0819|M.ulcerans_Agy99 IEPPCPACQSGDYSLCWSFTDGGIQPGIHTGVSANVTGGYAELMPAHDSM
MMAR_0254|M.marinum_M --------------------------------------------------
MLBr_01740|M.leprae_Br4923 --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAV_4597|M.avium_104 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4623|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_0819|M.ulcerans_Agy99 LFPVPGPISDEAAVFADPFSVSLHAITRHPPPRSGRVLVYGAGSLGLCAI
MMAR_0254|M.marinum_M --------------------------------------------------
MLBr_01740|M.leprae_Br4923 --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAV_4597|M.avium_104 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4623|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_0819|M.ulcerans_Agy99 AILRALYPDVAVAVVARFATQGELAGRFGAAKVVAHEPRLAIIEELVAWG
MMAR_0254|M.marinum_M --------------------------------------------------
MLBr_01740|M.leprae_Br4923 --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAV_4597|M.avium_104 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4623|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_0819|M.ulcerans_Agy99 GGRLRQALLGLPMAHPGAVDVVYDTVGKPETFEIGVRVLNSRGTLVKAGV
MMAR_0254|M.marinum_M --------------------------------------------------
MLBr_01740|M.leprae_Br4923 --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAV_4597|M.avium_104 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4623|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_0819|M.ulcerans_Agy99 HAPGRWEWSPLYFKEISWVGSNAFGVEEVEGVRKHAIAHYFDLVADDRLD
MMAR_0254|M.marinum_M --------------------------------------------------
MLBr_01740|M.leprae_Br4923 --------------------------------------------------
Mb1963c|M.bovis_AF2122/97 -------------------------------------
Rv1928c|M.tuberculosis_H37Rv -------------------------------------
MAB_4335|M.abscessus_ATCC_1997 -------------------------------------
TH_3427|M.thermoresistible__bu -------------------------------------
MAV_4597|M.avium_104 -------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 -------------------------------------
Mflv_0026|M.gilvum_PYR-GCK -------------------------------------
Mvan_4623|M.vanbaalenii_PYR-1 -------------------------------------
MUL_0819|M.ulcerans_Agy99 LRSMLTHTFALPQWRDAFLAIANQGESGAVKVAFDQR
MMAR_0254|M.marinum_M -------------------------------------
MLBr_01740|M.leprae_Br4923 -------------------------------------