For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAARREGALRRVAREIEAAGGRAMVAPLDVSSSE SVRAMVADVVGEFGRIDVVFNNAGVSLVGPVDAETFLDDTREMLEIDYLGTVRVVREVLPIMKQQRSGRI MNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQELRGSGIAVSVIHPALTQTPLLANVDPADMPPPFRS LTPIPVHWVAAAVLDGVARRRARVVVPFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYRGSVYRH QPTESAKAQAAQPERGYSSAR
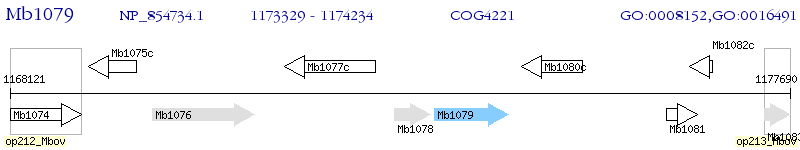
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1079 | - | - | 100% (301) | oxidoreductase |
| M. bovis AF2122 / 97 | Mb1570 | - | 4e-33 | 33.73% (255) | fatty acyl-CoA reductase |
| M. bovis AF2122 / 97 | Mb3423 | acrA1 | 7e-31 | 33.73% (255) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3370 | - | 2e-28 | 31.87% (251) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1050 | - | 1e-166 | 100.00% (301) | oxidoreductase |
| M. leprae Br4923 | MLBr_01094 | - | 4e-28 | 31.43% (280) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3919c | - | 3e-29 | 32.06% (262) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_2366 | - | 2e-35 | 34.40% (250) | fatty acyl-CoA reductase |
| M. avium 104 | MAV_3227 | - | 3e-36 | 35.60% (250) | oxidoreductase, short-chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_1623 | - | 1e-30 | 34.51% (255) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1828 | - | 1e-28 | 31.73% (249) | PROBABLE SHORT-CHAIN TYPE DEHYDROGENASE/REDUCTASE |
| M. ulcerans Agy99 | MUL_0918 | acrA1 | 8e-32 | 32.55% (255) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_3101 | - | 1e-31 | 33.47% (251) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3370|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3101|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
TH_1828|M.thermoresistible__bu --------------------------------------------------
MAB_3919c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2366|M.marinum_M --------------------------------------------------
MAV_3227|M.avium_104 --------------------------------------------------
MSMEG_1623|M.smegmatis_MC2_155 MRYVVTGGTGFIGRRVVSEILVRDASAEVWVLVRRESLNRFEHLALDWGE
MUL_0918|M.ulcerans_Agy99 MRYVVTGGTGFIGRRVVSRLLQSDADARVWVLVRRQSLSRFERLATDWGD
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3370|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3101|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
TH_1828|M.thermoresistible__bu --------------------------------------------------
MAB_3919c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2366|M.marinum_M --------------------------------------------------
MAV_3227|M.avium_104 --------------------------------------------------
MSMEG_1623|M.smegmatis_MC2_155 RAKPLVGDLRADDLGLTGADVADLGDVTHVVHCAAIYDITVDEAAQRAAN
MUL_0918|M.ulcerans_Agy99 RVKPLVGELP--ELPVTEQISAELGQIDHVVHCAAVYDITAGEDQQRAAN
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3370|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3101|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
TH_1828|M.thermoresistible__bu --------------------------------------------------
MAB_3919c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2366|M.marinum_M --------------------------------------------------
MAV_3227|M.avium_104 --------------------------------------------------
MSMEG_1623|M.smegmatis_MC2_155 VDGTRAVIALARRLEATLHHVSSIAVAGNHRGVFTEDDFDVAQDLPTPYH
MUL_0918|M.ulcerans_Agy99 VDGTRAVIELTRRLDATLHHVSSIAVAGDFAGEYTEADFDVGQQLPSPYH
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3370|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3101|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
TH_1828|M.thermoresistible__bu --------------------------------------------------
MAB_3919c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2366|M.marinum_M ------------------------------------------------MK
MAV_3227|M.avium_104 ------------------------------------------------MN
MSMEG_1623|M.smegmatis_MC2_155 QTKFEAEMLVRATTDVPHRIYRPAVVVGDSRTGEMDKIDGPYYFFGVLAR
MUL_0918|M.ulcerans_Agy99 RTKFEAELLVRSSTGLRYRIYRPAVVVGDSRTGEMDKIDGLYYFFGVLAR
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3370|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3101|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
TH_1828|M.thermoresistible__bu --------------------------------------------------
MAB_3919c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2366|M.marinum_M LGDLT------------------NLVEKPFAAMSNIVN------------
MAV_3227|M.avium_104 LGDLT------------------NLVEKPLAAVSNIIN------------
MSMEG_1623|M.smegmatis_MC2_155 LAALPRFTPMALPDTGRTNVVPVDFVAKALVHLMHVEG---ADGQTFHLT
MUL_0918|M.ulcerans_Agy99 LAALPSLTPILLPDTGRTNIVPVDYVVDALVALIHEDNMAFPNGRTFHLT
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3370|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3101|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
TH_1828|M.thermoresistible__bu --------------------------------------------------
MAB_3919c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2366|M.marinum_M TPNSAG--------------------------------RYRPFYLRNLLD
MAV_3227|M.avium_104 TPNSAG--------------------------------RYRPFYLRNLLD
MSMEG_1623|M.smegmatis_MC2_155 APNTVGLVDIYRGVAGAAGLPPLRAKLPRVTAAPVLDARGRARIVRNMVA
MUL_0918|M.ulcerans_Agy99 APQSVGLRGIYQAVTKEAGLPPARGVLPGSVVTPVLKARGRAKVLRNMAA
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3370|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3101|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
TH_1828|M.thermoresistible__bu --------------------------------------------------
MAB_3919c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2366|M.marinum_M VVQGRK--------------------------------------------
MAV_3227|M.avium_104 AVQGRT--------------------------------------------
MSMEG_1623|M.smegmatis_MC2_155 TQLGIPGEVFDVAELRPTFTADNTDTALRGTGIAVPEFASYAPQLFRYWA
MUL_0918|M.ulcerans_Agy99 TQLGIPSEIFDVLDLKPTFISDETQKALRGSGVRLPDFAGYAPKLWRYWA
Mb1079|M.bovis_AF2122/97 -----------MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAA
Rv1050|M.tuberculosis_H37Rv -----------MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAA
Mflv_3370|M.gilvum_PYR-GCK --------MASTDTAQLSGKVAFVTGGASGIGAALVAKLATAGTEVWIAD
Mvan_3101|M.vanbaalenii_PYR-1 -------------MAQLSGKVVFITGGASGIGAALTARLGAAGAQVWIAD
MLBr_01094|M.leprae_Br4923 -------------MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAISD
TH_1828|M.thermoresistible__bu -------------MQGFAGKVAVITGAGSGIGQALALELGRSGAKVAISD
MAB_3919c|M.abscessus_ATCC_199 -------------MKNFDNKVAVITGAGSGIGRSLALALAQRGARLALSD
MMAR_2366|M.marinum_M ------------LSDAVEGKVVLITGGSSGIGEAAAHKIAEAGGTVVLVA
MAV_3227|M.avium_104 ------------LEEAVDAKTVLITGASSGIGEAAAKKIAEAGGVVALVA
MSMEG_1623|M.smegmatis_MC2_155 EHLDPDRARRCDPAGPLVGRHVIITGASSGIGRASAIAVARRGAVVFALA
MUL_0918|M.ulcerans_Agy99 QHLDPDRALRDDPRGPLVGRHVIITGASSGIGRASALAVAQRGGCVFALA
. : ..:**..**** : . .. * :
Mb1079|M.bovis_AF2122/97 RREGALRRVAREIEAAGGRAMVAPLDVSSSESVRAMVADVVGEFGRIDVV
Rv1050|M.tuberculosis_H37Rv RREGALRRVAREIEAAGGRAMVAPLDVSSSESVRAMVADVVGEFGRIDVV
Mflv_3370|M.gilvum_PYR-GCK RQIAPAEELAAQLRATGAVAHAVELDVRDPAAFAHAVHTAVDTSGRLDYL
Mvan_3101|M.vanbaalenii_PYR-1 RQFGVAEELAQRLRDTGARVHAVELDVRDPAAFATAIAATVQTAGRIDYL
MLBr_01094|M.leprae_Br4923 VDGEGLAQTEGQLKAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQI
TH_1828|M.thermoresistible__bu VNTEGLTETEDRLKAIGAPVKADRLDVTERAAFEAYADAVVEHFGVVNQI
MAB_3919c|M.abscessus_ATCC_199 IDTAGVADTAGRCEKAGATAIPFELDVADRAAVYAHAADVKNEFGQVNLV
MMAR_2366|M.marinum_M RTRENLEKVADDVRAGGGAAHVYPCDLSDMDAIAAMADQVLADLGGVDIL
MAV_3227|M.avium_104 RTRENLEKVASEIRENGGSAHVYPCDLSDMDAIAAMADRVLDDLGGVDIL
MSMEG_1623|M.smegmatis_MC2_155 RNGEALDELIAEIRADGGQAHAFTCDVTDSASVEHTVKDILGRFDHVDYL
MUL_0918|M.ulcerans_Agy99 RNADALDQLVAEIRAGGGQAYAFTCDITDSTSVEHTVKDILGRFGHVDYL
. *. . *: . :. . :: :
Mb1079|M.bovis_AF2122/97 FNNAGVSLVGPVDAET-FLDDTREMLEIDYLGTVRVVREVLPIMKQQRSG
Rv1050|M.tuberculosis_H37Rv FNNAGVSLVGPVDAET-FLDDTREMLEIDYLGTVRVVREVLPIMKQQRSG
Mflv_3370|M.gilvum_PYR-GCK FNNAGIGIGGEVDTL--TLDDWTDIIDVNLKGVIHGVHAAYPQMIRQGSG
Mvan_3101|M.vanbaalenii_PYR-1 FNNAGIGIGGEVDTL--TLDDWNDIIDVNLRGVVHGIQAAYPIMIRQGSG
MLBr_01094|M.leprae_Br4923 YNNAGIAFTGDVEVS--HFKDIERVMDVDYWGVVNGTKAFLPYLISSGDG
TH_1828|M.thermoresistible__bu YNNAGIAYTGDIEVS--HFKDIERVMDVDFWGVVNGTKVFLPHLIASGDG
MAB_3919c|M.abscessus_ATCC_199 FNNAGVALSADVKDM--EWDDFDWLMNINFWGVAHGTKAFLPHLIESGDG
MMAR_2366|M.marinum_M INNAGRSIRRSLELSYDRIHDYQRTMQLNYLGAVQLILKFIPGMRERGFG
MAV_3227|M.avium_104 INNAGRSIRRSLELSYDRIHDYQRTMQLNYLGAVQLILKFIPGMRERGFG
MSMEG_1623|M.smegmatis_MC2_155 VNNAGRSIRRSVANSTDRLHDYERVMAVNYFGAVRMVLALLPHWRERRFG
MUL_0918|M.ulcerans_Agy99 VNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRMVLALLPHWRERRFG
**** . : .* : :: *. . * *
Mb1079|M.bovis_AF2122/97 RIMNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQELRGSG--IAVSVI
Rv1050|M.tuberculosis_H37Rv RIMNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQELRGSG--IAVSVI
Mflv_3370|M.gilvum_PYR-GCK HIVNTASMAGLITTSAQGGYSATKHAVVALSKTMRVEAATHG--VRVSVL
Mvan_3101|M.vanbaalenii_PYR-1 HIVNTASMAGLVTTSAQAGYSATKHAVVALSKTMRVEAATHG--VRVSVL
MLBr_01094|M.leprae_Br4923 HVINISSVFGLFSVPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTV
TH_1828|M.thermoresistible__bu HVINISSVFGLFSVPGQAAYNAAKFAVRGFTEALRQEMILAGHPVKVTTV
MAB_3919c|M.abscessus_ATCC_199 HIVNVSSVFGLMGIPSQSAYNAAKFAVRGFTESLRQEIRAAKFPVGVTCV
MMAR_2366|M.marinum_M HIINVSSVGVQTRAPRFGAYIASKAALDSLCDALQAETINEG--VRFTTV
MAV_3227|M.avium_104 QIINVSSVGVQTRAPRFGAYIASKAALDSLCDALQAEVVNDN--VKFTTV
MSMEG_1623|M.smegmatis_MC2_155 HVVNVSSAGVQAHTPKYSAYLPSKAALDAFSDVVSTETLSDH--ITFTNI
MUL_0918|M.ulcerans_Agy99 HVVNVSSAGVQARNPKYSSYLPSKAALDAFADVVASETLSDH--VTFTNI
:::* :* . ..* .: *: .: . : * : .: :
Mb1079|M.bovis_AF2122/97 HPALTQTPLLANVDPADMPPPFRSLTPIPVHWVAAAVLDGVARRRARVVV
Rv1050|M.tuberculosis_H37Rv HPALTQTPLLANVDPADMPPPFRSLTPIPVHWVAAAVLDGVARRRARVVV
Mflv_3370|M.gilvum_PYR-GCK CPGVVRTPILTGGAFGRNKSVSREDQLKLGEALRPMDAGVFAQKALRGVL
Mvan_3101|M.vanbaalenii_PYR-1 CPGVVRTPILSGGAYGRNKSVSREDLVKLGEALRPMDADDFADRALRAVL
MLBr_01094|M.leprae_Br4923 YPGGIKTAIARNAT-AAEGLDVSKIASRFDTWVAHTSPQHAARIILKAVR
TH_1828|M.thermoresistible__bu HPGGIKTGIARNMT-AAEGLDAQELARAFDTKLARTSPEKAAQIILDGVR
MAB_3919c|M.abscessus_ATCC_199 HPGGIKTNIASSARGIPDGVDRETVRKGFQ-LMAITRPDSAARIILRGVE
MMAR_2366|M.marinum_M HMALVRTPMISPTTIYDKFPTLTPEQAAGVITDAIVHRPRRASSPFGQFA
MAV_3227|M.avium_104 HMALVRTPMISPTTLYDKFPALTPEQAAGVIADAIVHRPRRASSPFGQFA
MSMEG_1623|M.smegmatis_MC2_155 HMPLVKTPMIAPSRRLNPVPPISAEHAAAMVVRALVEKPPRIDTPLGTFA
MUL_0918|M.ulcerans_Agy99 HMPLVATPMIVPSRRLNPTPAISAEHAAAMVVRGLVEKPARIDTPLGTLA
* : .
Mb1079|M.bovis_AF2122/97 PFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYRGSVYRHQPTESA
Rv1050|M.tuberculosis_H37Rv PFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYRGSVYRHQPTESA
Mflv_3370|M.gilvum_PYR-GCK RNEAIIVVP---RWWKALWYLERLSPALSMRTAAVALKR------TREVQ
Mvan_3101|M.vanbaalenii_PYR-1 RNEAIIVVP---RWWKALWYLERLSPALSMRTAAAALKR------TREVQ
MLBr_01094|M.leprae_Br4923 KKKARVLVG---PDAKVANVVVRFSGGAGYQRLFA------------QVA
TH_1828|M.thermoresistible__bu KNKARVLVG---TDAKILDAIVRLTG-SGYQRLFP------------KFV
MAB_3919c|M.abscessus_ATCC_199 KNRPRVLIG---PDARVFDAIPRIIG-PRYEDLMAPFYGMGRYGIGKKIA
MMAR_2366|M.marinum_M AVADAVNPAVMDRVRNRAFGMFSDSSAAKGSESQS-------------GT
MAV_3227|M.avium_104 AVADAVNPAVMDRVRNRAFAMFGDSSAAKGDESAA-------------DA
MSMEG_1623|M.smegmatis_MC2_155 DFGNYLTPRVARRFLHQLYLGYPDSAAARGLAPAEPRPASTAGPGLPERR
MUL_0918|M.ulcerans_Agy99 EIGNYVAPKLSRRILHQLYLGYPDSAAAQGISPLN-----DAQEHRPVRR
: .
Mb1079|M.bovis_AF2122/97 KAQAAQPERGYSSAR------------------
Rv1050|M.tuberculosis_H37Rv KAQAAQPERGYSSAR------------------
Mflv_3370|M.gilvum_PYR-GCK SAVPLQ---------------------------
Mvan_3101|M.vanbaalenii_PYR-1 SAVPPQ---------------------------
MLBr_01094|M.leprae_Br4923 SRLILNQR-------------------------
TH_1828|M.thermoresistible__bu ARTMPR---------------------------
MAB_3919c|M.abscessus_ATCC_199 GRFGIEL--------------------------
MMAR_2366|M.marinum_M TELDKRS--------------ETFVQATRGIHW
MAV_3227|M.avium_104 TEFDARS--------------ETFVRATRGIHW
MSMEG_1623|M.smegmatis_MC2_155 PKRPARSRRGVRVPRAVGRPVKRAVRTLPGVHW
MUL_0918|M.ulcerans_Agy99 PKRPALAVTGMRIP----RPVKRLVRLAPGVHW