For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSVVLPGDPIGSLIGDRRVTASSGGLHAHVYPATGLPNATVALAGAAEVDAAVSAAWEAHREWVAHPAD RRRDMLVGLADVVHEHLDELAGLNVHDYAVPISNAGTALLLERFLRHFAGYADKPHGTSSPVSGSFDVNI IEREPYGVVGVIAPWNGALAVAASCVAPALAAGNAVVFKPSELAPLAALRFGELCLVAGLPPGLVNILPA ASDGGDALVRHPGIRKIHFTGGGTTARAVLRAATDNLTPVVTELGGKSANIIFPDADLGRAAALAAHQGP LMQSGQSCACASRILVHESVYDAFTEQFLAVIGAARVGDPFDPEVNFGPVISAGSLDRLLNVVHRAVEER AGELLVGGERLVDGALAGGFYMQPTVFGGVDNRSRLAQVETFGPVVSLIRFVDDDEAVRLANDTPYGLNA FVHTADLTRAHTVSRQLEAGSVWVNQFSQISPQGPYGGYKQSGSGRTGGLDGLHEFQQIKNIRIGMG
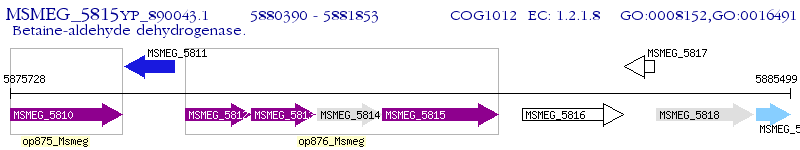
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5815 | - | - | 100% (487) | betaine aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6874 | - | 6e-82 | 37.26% (475) | aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6297 | - | 3e-80 | 36.67% (480) | aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2883c | aldC | 6e-76 | 39.87% (459) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1907 | - | 0.0 | 75.26% (481) | aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv2858c | aldC | 6e-76 | 39.87% (459) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 2e-49 | 28.98% (459) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0972 | - | 4e-74 | 37.29% (480) | putative aldehyde dehydrogenase |
| M. marinum M | MMAR_0780 | putA_1 | 1e-73 | 37.97% (482) | aldehyde dehydrogenase, PutA_1 |
| M. avium 104 | MAV_3553 | - | 0.0 | 66.81% (473) | aldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_3419 | - | 1e-116 | 48.00% (475) | PUTATIVE putative aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_1416 | - | 2e-74 | 38.17% (482) | aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0014 | - | 7e-78 | 39.22% (464) | betaine-aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5815|M.smegmatis_MC2_155 ---------MTSVVLPGDPIGSLIGDRRVTASSGGLHAHVYPATGLPNAT
Mflv_1907|M.gilvum_PYR-GCK -----------MVADPVTPIGLLIGTDRLTRTSLAPHPHIYPATGAPNAT
MAV_3553|M.avium_104 --------MTATASSPPRTGALLIGTERITAASGGSHQHRYPGTGVPNAT
TH_3419|M.thermoresistible__bu --------VQRTLPDKLPAPRLVIGGGDVISTSGGVYEHHYAATGGVQAE
Mb2883c|M.bovis_AF2122/97 -----------------------MSTTQLIN----------PATEEVLAS
Rv2858c|M.tuberculosis_H37Rv -----------------------MSTTQLIN----------PATEEVLAS
MMAR_0780|M.marinum_M MTVFARPGSAGALMSFESRYGNFIGGEWVAPARGRYFEDLTPVTGQPFCE
MUL_1416|M.ulcerans_Agy99 MTVFARPGSAGALMSFESRYGNLIGGEWVAPARGRYFEDLTPVTGQPFCE
MAB_0972|M.abscessus_ATCC_1997 MAKYAAPGTEGSAVTFAPRYDNFIGGEWVAPADGQYFENASPIDGKVFTR
Mvan_0014|M.vanbaalenii_PYR-1 -------------MSDIPHYRMYIDGAWRDAAD--TIEVRSPATGELVAT
MLBr_02573|M.leprae_Br4923 ----------------------------------MSIATINPATGETVKT
.
MSMEG_5815|M.smegmatis_MC2_155 VALAGAAEVDAAVSAAWEAHRE--WVAHPADRRRDMLVGLADVVHEHLDE
Mflv_1907|M.gilvum_PYR-GCK VALAGADEVGMAVSAAREAQRE--WITLGVDQRRDKLIDLADVVREHLGE
MAV_3553|M.avium_104 IPLAGHSEVDRAVHSAREAQRE--WISYPVDRRRDLLFGLADLLRDHFAE
TH_3419|M.thermoresistible__bu VPLAGPAEVDAAVAAARAAFPA--WRDLDLNRRRDILQELARRLVADGER
Mb2883c|M.bovis_AF2122/97 VDHTDANAVDDAVQRARAAQRR--WARLAPAQRAAGLRAFAAAVQAHLDE
Rv2858c|M.tuberculosis_H37Rv VDHTDANAVDDAVQRARAAQRR--WARLAPAQRAAGLRAFAAAVQAHLDE
MMAR_0780|M.marinum_M IPRSDEADVEMALDAAHAAAPA--WGKMAPAERAAILNKIADRIDENRAA
MUL_1416|M.ulcerans_Agy99 IPRSDEADVEMALDAAHAAAPA--WGKMAPAERAAILNKIADRIDENRAA
MAB_0972|M.abscessus_ATCC_1997 VARGTAPDIEKALDAAHAAAEA--WGNTSAAERSNALLRIADRMEQHIER
Mvan_0014|M.vanbaalenii_PYR-1 VAYGGVTTVDDAVAAAKAADEAGVWRNTSAQQRADVLDAIADNLAARTDE
MLBr_02573|M.leprae_Br4923 FTPTTQDEVEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEANQ
. : *: * : .* * :
MSMEG_5815|M.smegmatis_MC2_155 LAGLNVHDYAVP--ISNAGTALLLERFLRHFAGYADK---PHGTSSPVSG
Mflv_1907|M.gilvum_PYR-GCK LAELNTLDYGVP--ISYAGNAVMLDQFLRHFAGYVDK---PHGSSTPVSG
MAV_3553|M.avium_104 LTELNVHDYGVP--ISVAGNSVLTERFVRYYAGYPDK---LTGASTPVSG
TH_3419|M.thermoresistible__bu LGAIATLEVGTPN-LLASSSSAMAADWFTYYAGWIGR---SAGEVVPVPA
Mb2883c|M.bovis_AF2122/97 LAALEVANSGHPI-VSAEWEAGHVRDVLAFYAASPER---LSGRQIPVAG
Rv2858c|M.tuberculosis_H37Rv LAALEVANSGHPI-VSAEWEAGHVRDVLAFYAASPER---LSGRQIPVAG
MMAR_0780|M.marinum_M LAVAEVWDNGKPVREAIAADIPLAADHFRYFAAAIR----AQEGSLSQID
MUL_1416|M.ulcerans_Agy99 LAVAEVWDNGKPVREAIAADIPLAADHFRYFAAAIR----AQEGSLSQID
MAB_0972|M.abscessus_ATCC_1997 LAVAETWDNGKPIRESLNADIPLAIDHFRYFAGVLR----AQEGSISEID
Mvan_0014|M.vanbaalenii_PYR-1 LTALQVAENGATVRAAGAFLIGYAIAHLRYFAGLARTYAFQSSGPLMEAP
MLBr_02573|M.leprae_Br4923 SAALMTLEMGKTL-QSAKDEAMKCAKGFRYYAEKAET-LLADEPAEAAAV
. : . . . .:*
MSMEG_5815|M.smegmatis_MC2_155 SFDVNIIEREPYGVVGVIAPWNGALAVAASCVAPALAAGNAVVFKPSELA
Mflv_1907|M.gilvum_PYR-GCK SSDVNLIEREPFGAVGVIAPWNGSLVVAASCVAPALAAGNAVVFKPSELA
MAV_3553|M.avium_104 AFDVNIVEREPYGVVAVIAPWNGPLVVAGAAVAPALAAGNAVIFKPSELA
TH_3419|M.thermoresistible__bu GGALDYVLDEPLGVIAAIIPWNGPLISIGMKVAPALAAGNCVVLKPPENA
Mb2883c|M.bovis_AF2122/97 G--VDVTFNEPMGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAELT
Rv2858c|M.tuberculosis_H37Rv G--VDVTFNEPMGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAELT
MMAR_0780|M.marinum_M QDTVAYHFHEPLGVVGQIIPWNFPILMGAWKLAPALAAGNAVVLKPAEQT
MUL_1416|M.ulcerans_Agy99 QDTVAYQFHEPLGVVGQIIPWNFPILMGAWKLAPALAAGNAVVLKPAEQT
MAB_0972|M.abscessus_ATCC_1997 HDTVAYHFHEPLGVVGQIIPWNFPLLMAVWKLAPALAAGNCVVLKPAEQT
Mvan_0014|M.vanbaalenii_PYR-1 TLAAGMIVRDPVGVCAGMIPWNFPLLLAVWKLGPALAAGNTVVLKPDDQT
MLBr_02573|M.leprae_Br4923 GASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNV
:* *. : *** .: .*** *** :.* . .
MSMEG_5815|M.smegmatis_MC2_155 PLAALRFGELCLVAGLPPGLVNILPA-ASDGGDALVRHPGIRKIHFTGGG
Mflv_1907|M.gilvum_PYR-GCK PLAALRFGELCLEAGLPPGLVNILPA-AAEGGDALVRHAGIAKIHFTGGG
MAV_3553|M.avium_104 SLAPLRFGELCLEAGLPPGLVNVLPA-GPEGGEALVRHPGVGKVHFTGGH
TH_3419|M.thermoresistible__bu PFSALRFAELAAEAGLPPGVLNVIPG-GAEAGSALCSHPGVDKITFTGGG
Mb2883c|M.bovis_AF2122/97 PLTTMRLGELAVEAGLDEDLLQVLPGKGTVVGERFVTHPDIRKIVFTGST
Rv2858c|M.tuberculosis_H37Rv PLTTMRLGELAVEAGLDEDLLQVLPGKGTVVGERFVTHPDIRKIVFTGST
MMAR_0780|M.marinum_M PASVLYLMSLIGDL-LPPGVVNVVNGFGAEAGKPLASSNRIAKVAFTGET
MUL_1416|M.ulcerans_Agy99 PASVLYLMSLIGDL-LPPGVVNVVNGFGAEAGRPLASSNRIAKVAFTGET
MAB_0972|M.abscessus_ATCC_1997 PASILVLMELIADL-LPSGTVNVVNGFGVEAGKPLASSPRIAKIAFTGET
Mvan_0014|M.vanbaalenii_PYR-1 PLTLLELARAADEVGLPAGVLNVVTGAGPTVGARLAEHPDVRKVAFTGST
MLBr_02573|M.leprae_Br4923 PQCALYLADVIARGGFPEDCFQTLLI-SANGVEAILRDPRVAAATLTGSE
. : : : . .: : . : : :**
MSMEG_5815|M.smegmatis_MC2_155 TTARAVLRAATDNLTPVVTELGGKSANIIFP-----DADLGRAAALAAHQ
Mflv_1907|M.gilvum_PYR-GCK HTARAILRAAADNLTPVVTELGGKSANIIFD-----DADLDIAAVRSAHQ
MAV_3553|M.avium_104 ATAQQVLRAASDNVTPVTAELGGKSAYLVFA-----DADLDTAAVIAAHQ
TH_3419|M.thermoresistible__bu ETARKVFAAAAQALTPVVLELGGKSANIVFP-----DADLDAAATMAVSA
Mb2883c|M.bovis_AF2122/97 EVGKRVMAGAAAQVKRVTLELGGKSANIVFH-----DCDLERAATTAP-A
Rv2858c|M.tuberculosis_H37Rv EVGKRVMAGAAAQVKRVTLELGGKSANIVFH-----DCDLERAATTAP-A
MMAR_0780|M.marinum_M TTGRLIMQYASQNLIPVTLELGGKSPNIFFSDVMAKPDDFQDKALEG-FT
MUL_1416|M.ulcerans_Agy99 TTGRLIMQYASQNLIPVTLELGGKSPNIFFSDVMAKPDDFQDKALEG-FT
MAB_0972|M.abscessus_ATCC_1997 TTGRLIMQYATENIIPVTLELGGKSPNIFMPDVMAADDAFLDKALEG-FT
Mvan_0014|M.vanbaalenii_PYR-1 EVGKSVMRAAADTVKKVTLELGGKGASIVL------DDADLDLAVDGSLF
MLBr_02573|M.leprae_Br4923 PAGQAVGAIAGNEIKPTVLELGGSDPFIVMP------SADLDAAVATAVT
..: : * : .. ****... :.: *
MSMEG_5815|M.smegmatis_MC2_155 GPLMQSGQSCACASRILVHESVYDAFTEQFLAVIGAARVGDPFDPEVNFG
Mflv_1907|M.gilvum_PYR-GCK GPLMQSGQSCACASRILVHEAVYEAFTERFIDVVESARIGDPLDPATVFG
MAV_3553|M.avium_104 GPLTQAGQSCACASRILVEASVYVSFVEKLVAQVQAATIGDPLRPDVMLG
TH_3419|M.thermoresistible__bu GLMMLSGQGCILPTRLLVHDDVYDEMVTRVVAGADALTVGDPWQPTTMMG
Mb2883c|M.bovis_AF2122/97 GVFDNAGQDCCARSRILVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMG
Rv2858c|M.tuberculosis_H37Rv GVFDNAGQDCCARSRILVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMG
MMAR_0780|M.marinum_M MFALNQGEVCTCPSRSLIQADIYDEFLELAAIRTKAVRQGDPLNIETMLG
MUL_1416|M.ulcerans_Agy99 MFALNQGEVCTCPSRSLIQADIYDEFLELAAIRTKAVRQGDPLNIETMLG
MAB_0972|M.abscessus_ATCC_1997 MFALNQGEVCTCPSRALVHASIYDEFIARAITRVEAIVGGDPLDEDTMIG
Mvan_0014|M.vanbaalenii_PYR-1 AFLLMSGQACESGTRLLVHESIHDEFVRRMVARAETLVMGDPMSLASDLG
MLBr_02573|M.leprae_Br4923 GRVQNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVG
*: * .* : . :: : : *** .*
MSMEG_5815|M.smegmatis_MC2_155 PVISAGSLDRLLNVVHRAVEERAGELLVGGERLVDGALAGGFYMQPTVFG
Mflv_1907|M.gilvum_PYR-GCK PVISAAALDRIVGVVGEAVDSGSGDLLAGGRRLTEDSLAAGYYLEPTVLG
MAV_3553|M.avium_104 PVITEAAADRILGVIERAVAEKAGDLVTGGRRLG-GELASGYFIEPTIFS
TH_3419|M.thermoresistible__bu PLATAAQLERVTRAVDSALSDKAGKLLAGGRRPD--GLDAGYFYRPTVFG
Mb2883c|M.bovis_AF2122/97 PLVSRAHRDKVAGYVPDDAPVAFRGTAPAG---------RGFWFPPTVLT
Rv2858c|M.tuberculosis_H37Rv PLVSRAHRDKVAGYVPDDAPVAFRGTAPAG---------RGFWFPPTVLT
MMAR_0780|M.marinum_M SQASNDQLEKVLSYIEIGKEEGAKVITGGERAELGGDLSGGFYMQPTIFS
MUL_1416|M.ulcerans_Agy99 SQASNDQLEKVLSYIEIGKEEGAKVITGGERAELGGDLSGGFYMQPTIFS
MAB_0972|M.abscessus_ATCC_1997 AQASNDQYEKILSYIDIGRQEGAQVLTGGDARKVS-EYPDGAYIQPTVFA
Mvan_0014|M.vanbaalenii_PYR-1 PLVSAKQKARVEKYIALGQEEGCKLAYQG-TVPTDPALAQGHWVPPTILT
MLBr_02573|M.leprae_Br4923 PLATESGRAQVEQQVDAAAAAGAVIRCGGKRPDR-----PGWFYPPTVIT
. : :: : . * : **::
MSMEG_5815|M.smegmatis_MC2_155 GVDNRSRLAQVETFGPVVSLIRFVDDDEAVRLANDTPYGLNAFVHTADLT
Mflv_1907|M.gilvum_PYR-GCK NVDNTSPLAQTETFGPVVSLIPFADDDGAVRIANDTPYGLNAFVHTRDLT
MAV_3553|M.avium_104 NVDNRSSLAQTETFGPVASVMPFADEAQAVSLANDSRYGLNAFVATANLE
TH_3419|M.thermoresistible__bu DVDPASDLAQTEIFGPVLAILRFRSEEEAVELANNTQYGLAAYIHTRDLR
Mb2883c|M.bovis_AF2122/97 PK-RGDRTVTDEIFGPVVVVLTFDDEADAISLANDTAYGLSGSIWTDDLS
Rv2858c|M.tuberculosis_H37Rv PK-RGDRTVTDEIFGPVVVVLTFDDEADAISLANDTAYGLSGSIWTDDLS
MMAR_0780|M.marinum_M G-TNKMRIFQEEIFGPVVAVTPFTDYDDAMGIANDTLYGLGAGVWSRDGN
MUL_1416|M.ulcerans_Agy99 G-TNKMRIFQEEIFGPVVAVTPFTDYDDAMGIANDTLYGLGAGVWSRDGN
MAB_0972|M.abscessus_ATCC_1997 G-TNNMRIFQEEIFGPVLSVAKFDSLDEALKIANDTLYGLGAGVWSRDMT
Mvan_0014|M.vanbaalenii_PYR-1 GATNDMRIAREEIFGPVLVVLTYGDDDEAVAIANDSEYGLSAGVWSADRE
MLBr_02573|M.leprae_Br4923 DITKDMALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNES
* **** : . *: :** : :** . : :
MSMEG_5815|M.smegmatis_MC2_155 RAHTVSRQLEAGSVWVNQFSQISPQGPYGGYKQSGSGRTGGLDGLHEFQQ
Mflv_1907|M.gilvum_PYR-GCK RAHTVARRLQSGSVWINQGSRISPQGPYGGYKQSGSGRTGGLEGLREFQQ
MAV_3553|M.avium_104 RAHRVARRLQAGSVWVNRHSDIEPQGPYGGYKQSGFGRTGGVEGLHEFLQ
TH_3419|M.thermoresistible__bu RAHVMAAALDAGGVAVNGFPIVPSAAPFGGVKQSGFGREGGIWGLREFQR
Mb2883c|M.bovis_AF2122/97 RALRVARAVESGNLSVNSHSSVRFNTPFGGFKQSGVGRELGPDAPLQFTE
Rv2858c|M.tuberculosis_H37Rv RALRVARAVESGNLSVNSHSSVRFNTPFGGFKQSGVGRELGPDAPLQFTE
MMAR_0780|M.marinum_M TAYRAGRDIQAGRVWVNCYHVYPAHSAFGGYKQSGIGREGHQMMLGHYQQ
MUL_1416|M.ulcerans_Agy99 TAYRAGRDIQAGRVWVNCYHVYPAHSAFGGYKQSGIGREGHQKMLGHYQQ
MAB_0972|M.abscessus_ATCC_1997 NAYRLGRGIKAGRVWTNCYHQYPAHAAFGGYKKSGIGRENHKMMLDHYQQ
Mvan_0014|M.vanbaalenii_PYR-1 RALGIARRLQSGTVWVNDWHMINAMYPFGGVKQSGLGRELGPDALDEYTE
MLBr_02573|M.leprae_Br4923 EQRHFIDNIVAGQVFINGMTVSYPELPFGGIKRSGYGRELSTHGIREFCN
: :* : * .:** *:** ** .: .
MSMEG_5815|M.smegmatis_MC2_155 IKNIRIGMG------------------
Mflv_1907|M.gilvum_PYR-GCK VKNIRIGIG------------------
MAV_3553|M.avium_104 VKNIRIGMS------------------
TH_3419|M.thermoresistible__bu TKNVYIGLQ------------------
Mb2883c|M.bovis_AF2122/97 TKNVFIAVGEEM---------------
Rv2858c|M.tuberculosis_H37Rv TKNVFIAVGEEM---------------
MMAR_0780|M.marinum_M TKNLLVSYSDKALGLF-----------
MUL_1416|M.ulcerans_Agy99 TKNLLVSYSDKPLGLF-----------
MAB_0972|M.abscessus_ATCC_1997 TKNILVSYSPDAAGFF-----------
Mvan_0014|M.vanbaalenii_PYR-1 PKFIHVDMTDDRRKHVYPVVISAAAQG
MLBr_02573|M.leprae_Br4923 IKTVWIA--------------------
* : :