For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RRFFADTTPLRSPDFRRLWLAGIPTVIGANLTIFAVPVQLYALTQNSAYVGLAGLFALVPLVVFGLWGGA WADAMDRRLLLIIASVGLAVSSVLLWAQAALALNNVWLVLSMLSVQQAFYAINSPTRSAAIPRMVPGEQL PAANSLNMTVMQFGAIVGPLMAGLLLNWVDLSTLYLIDAITCLAPIWATFRLTPMPVGNAERGSARWGFG AVLEGFRYLAGNRVVLMSFVVDLIAMIFGMPRALFPQMAHESFGGPVEGGTVMALLAAAMSAGAVLGGVF SGWLPRVRRQGLAVVVAIVVWGVAMVGFGIAGGLAHGTAGLVLWIALAFLAVGGAADMVSAAFRSTILQQ AASDDLRGRLQGVFTVVVAGGPRLADTLHGAAAAAVGTTVAAAGGGVLVVVGVVIAALAVPAFVRYRVTV AEPRVSRDVESDSV*
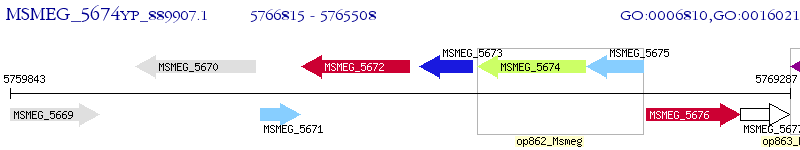
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5674 | - | - | 100% (435) | membrane transport protein |
| M. smegmatis MC2 155 | MSMEG_6390 | - | 8e-25 | 25.12% (430) | transporter, major facilitator family protein |
| M. smegmatis MC2 155 | MSMEG_0357 | - | 1e-21 | 26.00% (400) | transmembrane transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1288c | - | 1e-11 | 25.36% (418) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_1033 | - | 6e-23 | 25.87% (375) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv1258c | - | 1e-11 | 25.36% (418) | integral membrane transport protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0940 | - | 1e-162 | 72.43% (399) | hypothetical protein MAB_0940 |
| M. marinum M | MMAR_4182 | - | 1e-10 | 23.92% (393) | hypothetical protein MMAR_4182 |
| M. avium 104 | MAV_1406 | - | 1e-10 | 24.94% (393) | transporter, major facilitator family protein |
| M. thermoresistible (build 8) | TH_1317 | - | 1e-23 | 27.59% (395) | transmembrane transport protein |
| M. ulcerans Agy99 | MUL_0051 | - | 2e-06 | 25.07% (375) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_1167 | - | 3e-22 | 27.11% (402) | protein of unknown function DUF894, DitE |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5674|M.smegmatis_MC2_155 ------------MRRFFADTTPLRSPDFRRLWLAGIPTVIGANLTIFAVP
MAB_0940|M.abscessus_ATCC_1997 -----------------------------------MVTVIGAGLTLFAVP
TH_1317|M.thermoresistible__bu ------------------------------LWIAQFVSNLGTWMQTVGAQ
Mvan_1167|M.vanbaalenii_PYR-1 ----------MALAGSTSTWAPLGSPIFRALWIAQFVSNLGTWMQTVGAQ
Mb1288c|M.bovis_AF2122/97 -------------------MRNSNRGPAFLILFATLMAAAGDGVSIVAFP
Rv1258c|M.tuberculosis_H37Rv -------------------MRNSNRGPAFLILFATLMAAAGDGVSIVAFP
MMAR_4182|M.marinum_M -------------------MRNSSRGPAFLILFATLAACAGNGISIVAFP
MAV_1406|M.avium_104 -----------------------------MILFATLLATAGTGISIVAFP
Mflv_1033|M.gilvum_PYR-GCK MIQSPSRPAPVTSLPARGAFGLMFDRVFGALFWGKMFAVVAVWTHGLVAA
MUL_0051|M.ulcerans_Agy99 ---------MHASAPVELWRSVRSLPDFWRLLQVRIASQFGDGLFQAGLA
: : .
MSMEG_5674|M.smegmatis_MC2_155 VQLYALT---QNSAYVGLAGLFALVPLVVFGLWGGAWADAMD--RRLLLI
MAB_0940|M.abscessus_ATCC_1997 VQIYALT---QSSAYVGLTGVFGLVPLVVFGLWGGALADRMD--RRTLLI
TH_1317|M.thermoresistible__bu WMLVSTP---AAEVLVPLVQTATTLPIMLLSLPSGVVADLVD--RRRLLL
Mvan_1167|M.vanbaalenii_PYR-1 WMLVDDP---RAAVLVPLVQTATTLPVMLLALPSGVLADLID--RRRLLI
Mb1288c|M.bovis_AF2122/97 WLVLQRE---GSAGQASIVASATMLPLLFATLVAGTAVDYFG--RRRVSM
Rv1258c|M.tuberculosis_H37Rv WLVLQRE---GSAGQASIVASATMLPLLFATLVAGTAVDYFG--RRRVSM
MMAR_4182|M.marinum_M WLVLERE---GSAGQASIVAGAITLPLLFSTLVAGTAVDYFG--RRPVSM
MAV_1406|M.avium_104 WLALQHR---HSATDASIVAAAMTLPLVLATLIAGTAVDFFG--RRRVSL
Mflv_1033|M.gilvum_PYR-GCK IVMYDAT---RSALMVGLVGVAQFFPQLILAPTSGKWADTGNPARQILLG
MUL_0051|M.ulcerans_Agy99 GALLFNPDRAADPMAIARAFAVLFLPYSLLGPFAGALMDRWD--RRLVIV
. .* . .* * . *: :
MSMEG_5674|M.smegmatis_MC2_155 IASVGLAVSSVLLWAQAALALNNVWLVLSMLS---VQQAFYAINSPTRSA
MAB_0940|M.abscessus_ATCC_1997 ITATGLAASSVLLWLQAALSVDNVWVVLCLLS---VQQGFFAVNSPTRAA
TH_1317|M.thermoresistible__bu ATQSMMAAAVAILAALTAAQLTTPTVLLILLF---LIGCGQAFTAPAWQA
Mvan_1167|M.vanbaalenii_PYR-1 ATQGAMAAGVGLLAVLTGAGLTTPTVLLMLLF---VIGCGQALTAPAWQA
Mb1288c|M.bovis_AF2122/97 VAD-ALSGAAVAGVPLVAWGYGGDAVNVLVLA---VLAALAAAFGPAGMT
Rv1258c|M.tuberculosis_H37Rv VAD-ALSGAAVAGVPLVAWGYGGDAVNVLVLA---VLAALAAAFGPAGMT
MMAR_4182|M.marinum_M VSD-ALSGSAVAIVPLIAWAFGDDAVSVAVLA---ALGACSATFDPAGIT
MAV_1406|M.avium_104 VCD-WLSGAAVTAVPLTAWIFGAAAIDVAELA---VLAFCAAAFDPAGMT
Mflv_1033|M.gilvum_PYR-GCK RVFCVIGSGSIAVWMFAEPGLQGVAVAIPILIGTTLVGIGFVVGGPAMQS
MUL_0051|M.ulcerans_Agy99 GANIGRLILIAGIGTILAFEAGDLLLLFVALL----ANGLARFVASGLSA
: . * . :
MSMEG_5674|M.smegmatis_MC2_155 AIPRMVPGEQLPAANSLN------MTVMQFGAIVGP---LMAGLLLNWVD
MAB_0940|M.abscessus_ATCC_1997 AIPRMLPLEQLPAANALN------MTVQQFGFIAGP---LLAGVLLKWVD
TH_1317|M.thermoresistible__bu IQPELVPREQIPAAAALT------SMSLNAARAVGP---AVAGVLVAISG
Mvan_1167|M.vanbaalenii_PYR-1 IQPDLVPAHQIPAAAALG------SMSMNGARAIGP---AIAGALVSLTG
Mb1288c|M.bovis_AF2122/97 ARDSMLPEAAARAGWSLDRINGAYEAILNLAFIVGP---AIGGLMIATVG
Rv1258c|M.tuberculosis_H37Rv ARDSMLPEAAARAGWSLDRINGAYEAILNLAFIVGP---AIGGLMIATVG
MMAR_4182|M.marinum_M ARQSMLPEAAARAGWSLDRINSSYEAILNLAFVVGP---GIGGLMIATVG
MAV_1406|M.avium_104 ARQSMLPEAAARAGWSLDRTNSSYEAMLNLAFIVGP---GLGGLLIATLG
Mflv_1033|M.gilvum_PYR-GCK IVPTLIRNGELPTAMALN------SLPMTVGRVIGP---AAGAYLAAHVG
MUL_0051|M.ulcerans_Agy99 SLPHVVPREQVVTMNSVATASGAVAAFVGANFMLLPRWLAGGGDHGAAAV
:: : :: * ..
MSMEG_5674|M.smegmatis_MC2_155 LSTLYLIDAITCLAPIWATFRLTPMPVGNAER--GSARWGFGAVLEGFRY
MAB_0940|M.abscessus_ATCC_1997 LSTLYLIDAIACVAPIWATVRLHTMPPTGASKDLRANGSGLQDVLDGFRY
TH_1317|M.thermoresistible__bu PTLVFTLNAVSFAG---IVVVLLLWKRSSAEQTLAAER-PLAALQAGHRY
Mvan_1167|M.vanbaalenii_PYR-1 PTLVFALNAVSFIG---IVLVLISWRRPAVQRDFPPER-ALAALSAGGRY
Mb1288c|M.bovis_AF2122/97 GITTMWITATAFGLSILAIAALQLEGAGKPHHTSRPQG-LVSGIAEGLRF
Rv1258c|M.tuberculosis_H37Rv GITTMWITATAFGLSILAIAALQLEGAGKPHHTSRPQG-LVSGIAEGLRF
MMAR_4182|M.marinum_M GITTMWITAGAFGLSLLAIGALRLEGAGRPHHETRPDG-LVSGVTQGLRF
MAV_1406|M.avium_104 GINTMWVTAGCFALSFLAIGALRLEGAGKPPRATRPVG-LVTGIAEGVRF
Mflv_1033|M.gilvum_PYR-GCK PATAFTVSAVLHLV--FAMFLLAVRFPAPPQRRPGADY----RVRAAVRY
MUL_0051|M.ulcerans_Agy99 IFTAMIPVAISLLLALRFAPRVLGPDDTKRAIHGSAVYAVITGWMHGVRT
* : . . *
MSMEG_5674|M.smegmatis_MC2_155 LAGNRVVLMSFVVDLIAMIFG--MPRALFPQMAHESFGGPVEGGTVMALL
MAB_0940|M.abscessus_ATCC_1997 LAGHKIVLMSFVVDLVAMIFG--MPRILFPQIAHEGFGDPADGGTVIALL
TH_1317|M.thermoresistible__bu VRSSPVVRRILFRAALFILPAS-ALWGLLPVVASSGLGLSSSG---YGLM
Mvan_1167|M.vanbaalenii_PYR-1 IRSSPIVRRILLRTVLFIAPAS-ALWGLLPVIAANQLGLGSAG---YGLL
Mb1288c|M.bovis_AF2122/97 VWNLRVLRTLGMIDLTVTALYLPMESVLFPKYFTDHQQPVQLG-----WA
Rv1258c|M.tuberculosis_H37Rv VWNLRVLRTLGMIDLTVTALYLPMESVLFPKYFTDHQQPVQLG-----WA
MMAR_4182|M.marinum_M VWNLRVLRALALIDLTVTALYLPMESVLFPTYFSDRQQPAQLG-----SV
MAV_1406|M.avium_104 VWNLRVLRTLGLIDLAVTALYLPMESVLFPKYFADQHQPAQLG-----WA
Mflv_1033|M.gilvum_PYR-GCK VLADRPLLLALLAVTTVGIVSDPSITLAPSMADELGGGAAMVG-----TL
MUL_0051|M.ulcerans_Agy99 VVQIPTVAAALSGLAAHRMVVG-VNSLLILLLVHHMRDVDVEGLGTALLF
: : *
MSMEG_5674|M.smegmatis_MC2_155 AAAMSAGAVLGGVFSGWLPRVRRQGLAVVVAIVVWGVAMVGFGIAGGLAH
MAB_0940|M.abscessus_ATCC_1997 SAAISVGAVIGGVFSGWFQRIDRQGLAVVVSIVVWGLAMVGFGLT---AH
TH_1317|M.thermoresistible__bu LGALGAGAVLGALTLSAWQSAFRQNTLLALAAVGFAAATAVAVLV-----
Mvan_1167|M.vanbaalenii_PYR-1 LGALGAGAVLGAFVLSGLRTRFGQNVLLTAGAAGFAVATAVLALV-----
Mb1288c|M.bovis_AF2122/97 LMAIAGGGLVGALGYAVLAIRVPRRVTMSTAVLTLGLASMVIAFL-----
Rv1258c|M.tuberculosis_H37Rv LMAIAGGGLVGALGYAVLAIRVPRRVTMSTAVLTLGLASMVIAFL-----
MMAR_4182|M.marinum_M LVALSLGSLVGALGYGMLSNYVSRRTIMLTAVLTLGAATTGIALL-----
MAV_1406|M.avium_104 LMALALGGVAGALGYAVLSARLRRRTAVLTATLTFGATTAGIAVL-----
Mflv_1033|M.gilvum_PYR-GCK SAVFGVGAALGMGSLAVLRGRIASAYVSSIGMVALTLGSAVLAVG-----
MUL_0051|M.ulcerans_Agy99 FAATGLGAFLANVLTPGLIRRWG-RYATTNGALAAAAVTQLAGAG-----
. . *. . .
MSMEG_5674|M.smegmatis_MC2_155 GTAGLVLWIALAFLAVGGAADMVSAAFRSTILQQAASDDLRGRLQGVFTV
MAB_0940|M.abscessus_ATCC_1997 ---APLLWLALLFLVIGGIADMVSAAFRTTILQSVATDDVRGRLQGVFTV
TH_1317|M.thermoresistible__bu ----PDVVVVLIGLTVGGMSWLLALATLNASMQLSLPAWVRARGLSVYLL
Mvan_1167|M.vanbaalenii_PYR-1 ----PDFAAVLAALVVGGGAWLLSLSTLNASMQLSLPAWVRARGLSVYQL
Mb1288c|M.bovis_AF2122/97 ----PPLPVIMVLCAVVGLVYGPIQPIYNYVIQTRAAQHLRGRVVGVMTS
Rv1258c|M.tuberculosis_H37Rv ----PPLPVIMVLCAVVGLVYGPIQPIYNYVIQTRAAQHLRGRVVGVMTS
MMAR_4182|M.marinum_M ----PPLPVILVLCALIGVVYGPIQPIYNYVMQTRAPHYLRGRVVGVMTS
MAV_1406|M.avium_104 ----PPLPVILGLCAVTGVVYGPIQPIYNFVMQTRAPHHLRGRVVGVMAG
Mflv_1033|M.gilvum_PYR-GCK ----MQSWIAYTGFALAGFGFGCAMTGLSTVVQERAPEELRGRIMALWLV
MUL_0051|M.ulcerans_Agy99 ----LLLPVMVVCGFLLGVAGQVVKLCADSAMQIDVDGALRGHVFAVQDA
: * :* :*.: .:
MSMEG_5674|M.smegmatis_MC2_155 VVAGGPRLADTLHGAAAAAVGTTVAAAGGGVLVVVG--------------
MAB_0940|M.abscessus_ATCC_1997 VVAGGPRVADFAHGAAAAAVGTTAAAAGGGALVVVG--------------
TH_1317|M.thermoresistible__bu IFMGGQALGSTMWGLVAGSTSTVVAMLISAGLLVVSGLSAWWWPLHPQTG
Mvan_1167|M.vanbaalenii_PYR-1 IFMGGQAIGSLLWGLLAGATNSVTSLLVSAGLLVVCALSSLWWPLHAGTG
Mb1288c|M.bovis_AF2122/97 LAYAAGPLGLLLAGPLTDAAGLHATFLALALPIVCTG-------------
Rv1258c|M.tuberculosis_H37Rv LAYAAGPLGLLLAGPLTDAAGLHATFLALALPIVCTG-------------
MMAR_4182|M.marinum_M LAYAAGPLGLLLAGPVADAAGLKVAFFALALPIAITG-------------
MAV_1406|M.avium_104 LTYAAGPLGLLVAGPLADAAGLKATFLTLAVPILAIG-------------
Mflv_1033|M.gilvum_PYR-GCK GFLGSRPIAAAVLGGTADLTNVFVAFAVASGLALVVT-------------
MUL_0051|M.ulcerans_Agy99 LFWVSYIVSVTVAAALIPDDGRAPVFVLVGSLIYLLG-------------
. :. . . .
MSMEG_5674|M.smegmatis_MC2_155 ----------------------------------------VVIAALAVPA
MAB_0940|M.abscessus_ATCC_1997 ----------------------------------------VIVAALLVPV
TH_1317|M.thermoresistible__bu QLDVSPSAHWPEPALVFEPEPRDGPVLVLSTYRVAPEDEPAFLVAMQRQA
Mvan_1167|M.vanbaalenii_PYR-1 NLDLTPSSHWPEPTLVFEPEPLDGPVLVLTSYCVPAHNEGAFLAAMAVLG
Mb1288c|M.bovis_AF2122/97 ------------------------------------------LVAIRLPA
Rv1258c|M.tuberculosis_H37Rv ------------------------------------------LVAIRLPA
MMAR_4182|M.marinum_M ------------------------------------------LMCIRLPS
MAV_1406|M.avium_104 ------------------------------------------VVACGLPS
Mflv_1033|M.gilvum_PYR-GCK -------------------------------------------VLCRPSK
MUL_0051|M.ulcerans_Agy99 ---------------------------------------------LCVHS
MSMEG_5674|M.smegmatis_MC2_155 FVRYRVTVAEPRVSRDVESDSV----------------------------
MAB_0940|M.abscessus_ATCC_1997 FVRFRT-------STEAQLDPQ----------------------------
TH_1317|M.thermoresistible__bu RSRQRTGAALWRLYRHLEREHTFVEAFLVRSWDEHLRQHHVRLTLVDQRV
Mvan_1167|M.vanbaalenii_PYR-1 RSRQRTGASQWRLFRSVEQESTFVETFIVRSWGEHMHQHYTRLTGQDQLI
Mb1288c|M.bovis_AF2122/97 LRELDLAPQADIDRPVGSAQ------------------------------
Rv1258c|M.tuberculosis_H37Rv LRELDLAPQADIDRPVGSAQ------------------------------
MMAR_4182|M.marinum_M LRELDHAPTQDISETSLL--------------------------------
MAV_1406|M.avium_104 LRELDRAPQFADDPGA----------------------------------
Mflv_1033|M.gilvum_PYR-GCK LVDLR---------------------------------------------
MUL_0051|M.ulcerans_Agy99 IVGRRSSASNVEGC------------------------------------
MSMEG_5674|M.smegmatis_MC2_155 ------------------------------
MAB_0940|M.abscessus_ATCC_1997 ------------------------------
TH_1317|M.thermoresistible__bu EEEVRRYAEGPPLVEHLIAVNFDEITRRGV
Mvan_1167|M.vanbaalenii_PYR-1 EQNVEQYTDGTPVSQHYLAV-------RDV
Mb1288c|M.bovis_AF2122/97 ------------------------------
Rv1258c|M.tuberculosis_H37Rv ------------------------------
MMAR_4182|M.marinum_M ------------------------------
MAV_1406|M.avium_104 ------------------------------
Mflv_1033|M.gilvum_PYR-GCK ------------------------------
MUL_0051|M.ulcerans_Agy99 ------------------------------