For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTATPAMGATSTWAPLASPIYRALWIAQFVSNLGTWMQTVGAQWMLVGDPRAAVLVPLVQTATTLPVML LALPSGVLADLVDRRRLLIAAQGATAAGVASLATLTGAGLATPTVLLTLLFLIGCGQALTAPAWQAIQPD LVPREQIPAAAALGSMSMNGARAIGPAIAGALVSLSGPTLVFALNAVSFVGIVAVLLLWRRPAAERMLPA ERPLAALSAGGRYIRSAPIVRRILLRTVLFIAPGSALWGLLAVIAKDQLDLTSSGYGVLLGALGLGAVCG AFVLGRLQKMFELNTLLVVAAIGFGGATVVLALVHQVAVVVIALVLGGTAWLLALSTLNASMQLSLPGWV RARGLSVYQLVFMGGQALGSLLWGLIAGAAGSVTALLVSAGLMGVCAVSAIWWPLHARTGNLDMTPSAHW PEPALIFEPEPLDGPVVVLVSYRVEPAQETGFFEAMAALGRSRQRTGASQWQLFRSGEHAGTFVEAFIVR SWEEHLRQHHTRLTGHDLLVEETVRRFTVGEPTSQHLIAAKTH
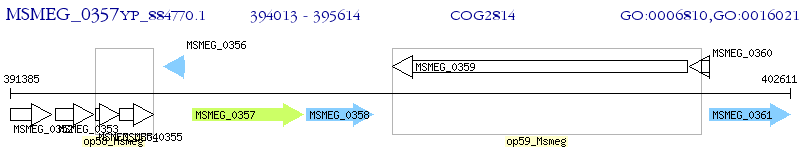
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0357 | - | - | 100% (533) | transmembrane transport protein |
| M. smegmatis MC2 155 | MSMEG_6390 | - | 3e-26 | 26.90% (394) | transporter, major facilitator family protein |
| M. smegmatis MC2 155 | MSMEG_1329 | - | 5e-25 | 25.83% (391) | major facilitator superfamily protein MFS_1, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1288c | - | 1e-14 | 21.76% (409) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_5163 | - | 0.0 | 77.59% (522) | protein of unknown function DUF894, DitE |
| M. tuberculosis H37Rv | Rv1258c | - | 1e-14 | 21.76% (409) | integral membrane transport protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0880 | - | 1e-44 | 33.88% (425) | hypothetical protein MAB_0880 |
| M. marinum M | MMAR_4182 | - | 6e-19 | 23.21% (405) | hypothetical protein MMAR_4182 |
| M. avium 104 | MAV_1406 | - | 6e-17 | 23.38% (385) | transporter, major facilitator family protein |
| M. thermoresistible (build 8) | TH_1317 | - | 0.0 | 70.30% (505) | transmembrane transport protein |
| M. ulcerans Agy99 | MUL_1084 | - | 1e-06 | 24.30% (391) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_1167 | - | 0.0 | 78.01% (523) | protein of unknown function DUF894, DitE |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1288c|M.bovis_AF2122/97 -------------MRNSNRGPAFLILFATLMAAAGDGVSIVAFPWLVLQR
Rv1258c|M.tuberculosis_H37Rv -------------MRNSNRGPAFLILFATLMAAAGDGVSIVAFPWLVLQR
MMAR_4182|M.marinum_M -------------MRNSSRGPAFLILFATLAACAGNGISIVAFPWLVLER
MAV_1406|M.avium_104 -----------------------MILFATLLATAGTGISIVAFPWLALQH
Mflv_5163|M.gilvum_PYR-GCK MTSP----VSTSTWAPLQSPVFRALWIAQFVSNLGTWMQTVGAQWMLVDD
Mvan_1167|M.vanbaalenii_PYR-1 MALA----GSTSTWAPLGSPIFRALWIAQFVSNLGTWMQTVGAQWMLVDD
MSMEG_0357|M.smegmatis_MC2_155 MTTATPAMGATSTWAPLASPIYRALWIAQFVSNLGTWMQTVGAQWMLVGD
TH_1317|M.thermoresistible__bu ------------------------LWIAQFVSNLGTWMQTVGAQWMLVST
MAB_0880|M.abscessus_ATCC_1997 ------MADRGGIFVSLRTRNYRLWISGQMVSLIGTWMQRVAQDWLVLDL
MUL_1084|M.ulcerans_Agy99 MTSEARTATTAARPWTPRVAAQLSVLAAAAFIYVTAEILPVGALSAIARN
. : *.
Mb1288c|M.bovis_AF2122/97 EG-SAGQASIVASATMLPLLFATLVAGTAVDYFGRRRVSMVADALSGAAV
Rv1258c|M.tuberculosis_H37Rv EG-SAGQASIVASATMLPLLFATLVAGTAVDYFGRRRVSMVADALSGAAV
MMAR_4182|M.marinum_M EG-SAGQASIVAGAITLPLLFSTLVAGTAVDYFGRRPVSMVSDALSGSAV
MAV_1406|M.avium_104 RH-SATDASIVAAAMTLPLVLATLIAGTAVDFFGRRRVSLVCDWLSGAAV
Mflv_5163|M.gilvum_PYR-GCK PA-AAVLVPLVQTATTLPVMLLALPSGVLADLVDRRRLLIATQGAMAAGV
Mvan_1167|M.vanbaalenii_PYR-1 PR-AAVLVPLVQTATTLPVMLLALPSGVLADLIDRRRLLIATQGAMAAGV
MSMEG_0357|M.smegmatis_MC2_155 PR-AAVLVPLVQTATTLPVMLLALPSGVLADLVDRRRLLIAAQGATAAGV
TH_1317|M.thermoresistible__bu PA-AEVLVPLVQTATTLPIMLLSLPSGVVADLVDRRRLLLATQSMMAAAV
MAB_0880|M.abscessus_ATCC_1997 SHGNAFLLGVVMALQFGPTLLLSVWGGMLADRYDKRRMLMLTQALMALFA
MUL_1084|M.ulcerans_Agy99 LHISVVLVGTLLSWYALVAALTTVPLVRWTAHWPRRHALMVS-----LVC
: : :: . :* :
Mb1288c|M.bovis_AF2122/97 AGVP-LVAWGYGGDAVNVLVLAVLAALAAAFGPAGMTARDSMLPEAAARA
Rv1258c|M.tuberculosis_H37Rv AGVP-LVAWGYGGDAVNVLVLAVLAALAAAFGPAGMTARDSMLPEAAARA
MMAR_4182|M.marinum_M AIVP-LIAWAFGDDAVSVAVLAALGACSATFDPAGITARQSMLPEAAARA
MAV_1406|M.avium_104 TAVP-LTAWIFGAAAIDVAELAVLAFCAAAFDPAGMTARQSMLPEAAARA
Mflv_5163|M.gilvum_PYR-GCK GLLATLTGAGLTTPTVLLTLLFVIGCGQALTAPAWQAIQPDLVPSDQIPA
Mvan_1167|M.vanbaalenii_PYR-1 GLLAVLTGAGLTTPTVLLMLLFVIGCGQALTAPAWQAIQPDLVPAHQIPA
MSMEG_0357|M.smegmatis_MC2_155 ASLATLTGAGLATPTVLLTLLFLIGCGQALTAPAWQAIQPDLVPREQIPA
TH_1317|M.thermoresistible__bu AILAALTAAQLTTPTVLLILLFLIGCGQAFTAPAWQAIQPELVPREQIPA
MAB_0880|M.abscessus_ATCC_1997 LTVGLLQVTGLVRIWHVMAVAFAMGCAAAIEAPTRQSFTIEMVGPEHLPN
MUL_1084|M.ulcerans_Agy99 LTISQLISALAPNFAVLAAGRALCAITHGLLWSVIAPIATRLVPPSHAGR
: * . . .. . ::
Mb1288c|M.bovis_AF2122/97 GWSLDRINGAYEAILNLAFIVGPAIGGLMIATVGGITTMWITATAFGLSI
Rv1258c|M.tuberculosis_H37Rv GWSLDRINGAYEAILNLAFIVGPAIGGLMIATVGGITTMWITATAFGLSI
MMAR_4182|M.marinum_M GWSLDRINSSYEAILNLAFVVGPGIGGLMIATVGGITTMWITAGAFGLSL
MAV_1406|M.avium_104 GWSLDRTNSSYEAMLNLAFIVGPGLGGLLIATLGGINTMWVTAGCFALSF
Mflv_5163|M.gilvum_PYR-GCK AAALGSMS------MNGARAIGPAIAGALVSLTGPTIVFALNAVSFVGIV
Mvan_1167|M.vanbaalenii_PYR-1 AAALGSMS------MNGARAIGPAIAGALVSLTGPTLVFALNAVSFIGIV
MSMEG_0357|M.smegmatis_MC2_155 AAALGSMS------MNGARAIGPAIAGALVSLSGPTLVFALNAVSFVGIV
TH_1317|M.thermoresistible__bu AAALTSMS------LNAARAVGPAVAGVLVAISGPTLVFTLNAVSFAGIV
MAB_0880|M.abscessus_ATCC_1997 AVGLNSMV------FNLAGIIGPATSGVLIGLIGTGWVFLLNFVSFIAVG
MUL_1084|M.ulcerans_Agy99 ATTSIYIG------TSLALVIGSPLTAALSLMWGWRLAAVCVTVAAAVVT
. . * :*. . : * . .
Mb1288c|M.bovis_AF2122/97 LAIAALQLEGAGKPHHTSRPQGLVSGIAEGLRFVWNLRVLRTLGMIDLTV
Rv1258c|M.tuberculosis_H37Rv LAIAALQLEGAGKPHHTSRPQGLVSGIAEGLRFVWNLRVLRTLGMIDLTV
MMAR_4182|M.marinum_M LAIGALRLEGAGRPHHETRPDGLVSGVTQGLRFVWNLRVLRALALIDLTV
MAV_1406|M.avium_104 LAIGALRLEGAGKPPRATRPVGLVTGIAEGVRFVWNLRVLRTLGLIDLAV
Mflv_5163|M.gilvum_PYR-GCK LVLMWWRRPPVRSDYPPERA---LAALSAGGRYIRSSPIVRRILLRTVLF
Mvan_1167|M.vanbaalenii_PYR-1 LVLISWRRPAVQRDFPPERA---LAALSAGGRYIRSSPIVRRILLRTVLF
MSMEG_0357|M.smegmatis_MC2_155 AVLLLWRRPAAERMLPAERP---LAALSAGGRYIRSAPIVRRILLRTVLF
TH_1317|M.thermoresistible__bu VVLLLWKRSSAEQTLAAERP---LAALQAGHRYVRSSPVVRRILFRAALF
MAB_0880|M.abscessus_ATCC_1997 IALWRMRPGELHSSEPIPAAKG---QLRQGFSYVWRRRDLFMIMLTVFLV
MUL_1084|M.ulcerans_Agy99 VAARLLLPEMVLSADQ-------LQHVGPRSRHHRN-RALIIVSLITMVG
. : . : : :
Mb1288c|M.bovis_AF2122/97 TALYLPMESVLFPKYFTDH-QQPVQLGWALMAIAGGGLVGALGYAV-LAI
Rv1258c|M.tuberculosis_H37Rv TALYLPMESVLFPKYFTDH-QQPVQLGWALMAIAGGGLVGALGYAV-LAI
MMAR_4182|M.marinum_M TALYLPMESVLFPTYFSDR-QQPAQLGSVLVALSLGSLVGALGYGM-LSN
MAV_1406|M.avium_104 TALYLPMESVLFPKYFADQ-HQPAQLGWALMALALGGVAGALGYAV-LSA
Mflv_5163|M.gilvum_PYR-GCK IAPGSAVWGLLPVIARDQLGLGSAGYGALLGALGVGAVLGAFALSR-LRA
Mvan_1167|M.vanbaalenii_PYR-1 IAPASALWGLLPVIAANQLGLGSAGYGLLLGALGAGAVLGAFVLSG-LRT
MSMEG_0357|M.smegmatis_MC2_155 IAPGSALWGLLAVIAKDQLDLTSSGYGVLLGALGLGAVCGAFVLGR-LQK
TH_1317|M.thermoresistible__bu ILPASALWGLLPVVASSGLGLSSSGYGLMLGALGAGAVLGALTLSA-WQS
MAB_0880|M.abscessus_ATCC_1997 STFGLNFSLTLAVLARETFGRGAESYGLLSTALAIGTLLGATFAAR-RTK
MUL_1084|M.ulcerans_Agy99 VTGHFVSYTYIVVIIRQVVGVRGPSLAWLLAAYGVAGVVAVALVARPLDR
: . * . . : .. .
Mb1288c|M.bovis_AF2122/97 RVPRRVTMSTAVLTLGLASMVIAFLPPLPVIMVLCAVVGLVYGPIQPIYN
Rv1258c|M.tuberculosis_H37Rv RVPRRVTMSTAVLTLGLASMVIAFLPPLPVIMVLCAVVGLVYGPIQPIYN
MMAR_4182|M.marinum_M YVSRRTIMLTAVLTLGAATTGIALLPPLPVILVLCALIGVVYGPIQPIYN
MAV_1406|M.avium_104 RLRRRTAVLTATLTFGATTAGIAVLPPLPVILGLCAVTGVVYGPIQPIYN
Mflv_5163|M.gilvum_PYR-GCK RFGQNKLLIAGAAGFGMATAVLALVHNVALVAAALVVGGAAWLLTLSTLN
Mvan_1167|M.vanbaalenii_PYR-1 RFGQNVLLTAGAAGFAVATAVLALVPDFAAVLAALVVGGGAWLLSLSTLN
MSMEG_0357|M.smegmatis_MC2_155 MFELNTLLVVAAIGFGGATVVLALVHQVAVVVIALVLGGTAWLLALSTLN
TH_1317|M.thermoresistible__bu AFRQNTLLALAAVGFAAATAVAVLVPDVVVVLIGLTVGGMSWLLALATLN
MAB_0880|M.abscessus_ATCC_1997 KVR-MTLFYGAAMAFGLSEVVVSVMPTYLSVALALVPAGLLALTFTTSAM
MUL_1084|M.ulcerans_Agy99 RPKGTIIFCVAGLTFAFVLLTALAFGGHLAPMTALVVGTGAIVLWGAAVT
. . :. . . .
Mb1288c|M.bovis_AF2122/97 YVIQTRAAQHLRG------RVVGVMTSLAYAAGPLGLLLAGPLTDAAGLH
Rv1258c|M.tuberculosis_H37Rv YVIQTRAAQHLRG------RVVGVMTSLAYAAGPLGLLLAGPLTDAAGLH
MMAR_4182|M.marinum_M YVMQTRAPHYLRG------RVVGVMTSLAYAAGPLGLLLAGPVADAAGLK
MAV_1406|M.avium_104 FVMQTRAPHHLRG------RVVGVMAGLTYAAGPLGLLVAGPLADAAGLK
Mflv_5163|M.gilvum_PYR-GCK ASMQLSLPSWVRA------RGLSVYQLIFMGGQAVGSLLWGLLAGGTSSV
Mvan_1167|M.vanbaalenii_PYR-1 ASMQLSLPAWVRA------RGLSVYQLIFMGGQAIGSLLWGLLAGATNSV
MSMEG_0357|M.smegmatis_MC2_155 ASMQLSLPGWVRA------RGLSVYQLVFMGGQALGSLLWGLIAGAAGSV
TH_1317|M.thermoresistible__bu ASMQLSLPAWVRA------RGLSVYLLIFMGGQALGSTMWGLVAGSTSTV
MAB_0880|M.abscessus_ATCC_1997 NIIQLSVDSQLRG------RVMGIYMVAFLGGTPLGSPLVGWLADLHGGR
MUL_1084|M.ulcerans_Agy99 AVSPMLQSAAMRSGADDPDGASGLYVTAFQVGIMAGSLIGGLLYERS--V
:*. .: . * : * :
Mb1288c|M.bovis_AF2122/97 ATFLALALPIVCTGLVAIRLPALR---ELDLAPQADIDRPVGSAQ-----
Rv1258c|M.tuberculosis_H37Rv ATFLALALPIVCTGLVAIRLPALR---ELDLAPQADIDRPVGSAQ-----
MMAR_4182|M.marinum_M VAFFALALPIAITGLMCIRLPSLR---ELDHAPTQDISETSLL-------
MAV_1406|M.avium_104 ATFLTLAVPILAIGVVACGLPSLR---ELDRAPQFADDPGA---------
Mflv_5163|M.gilvum_PYR-GCK ISLLVSAGLLIFCGLSLWWWPLHAGTGNLDLTPSSHWPEPTLLFEPEPLD
Mvan_1167|M.vanbaalenii_PYR-1 TSLLVSAGLLVVCALSSLWWPLHAGTGNLDLTPSSHWPEPTLVFEPEPLD
MSMEG_0357|M.smegmatis_MC2_155 TALLVSAGLMGVCAVSAIWWPLHARTGNLDMTPSAHWPEPALIFEPEPLD
TH_1317|M.thermoresistible__bu VAMLISAGLLVVSGLSAWWWPLHPQTGQLDVSPSAHWPEPALVFEPEPRD
MAB_0880|M.abscessus_ATCC_1997 MPLLVGGLVSAAAGLVCATYLHRHP----CPVVAGEVPEADDMVPPAPAA
MUL_1084|M.ulcerans_Agy99 AMMLTASAGLMGVALVGMTSARRVFEAPVAVAGATSHNQ-----------
:: . .:
Mb1288c|M.bovis_AF2122/97 --------------------------------------------------
Rv1258c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4182|M.marinum_M --------------------------------------------------
MAV_1406|M.avium_104 --------------------------------------------------
Mflv_5163|M.gilvum_PYR-GCK GPVLVITAYRVLPENEEPFMSAMARLGRSRQRTGASMWQLFRSIEQESTF
Mvan_1167|M.vanbaalenii_PYR-1 GPVLVLTSYCVPAHNEGAFLAAMAVLGRSRQRTGASQWRLFRSVEQESTF
MSMEG_0357|M.smegmatis_MC2_155 GPVVVLVSYRVEPAQETGFFEAMAALGRSRQRTGASQWQLFRSGEHAGTF
TH_1317|M.thermoresistible__bu GPVLVLSTYRVAPEDEPAFLVAMQRQARSRQRTGAALWRLYRHLEREHTF
MAB_0880|M.abscessus_ATCC_1997 TPVCEPTPLARRS-------------------------------------
MUL_1084|M.ulcerans_Agy99 --------------------------------------------------
Mb1288c|M.bovis_AF2122/97 --------------------------------------------------
Rv1258c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4182|M.marinum_M --------------------------------------------------
MAV_1406|M.avium_104 --------------------------------------------------
Mflv_5163|M.gilvum_PYR-GCK VETFIVRSWGEHMHQHYTRLTGQDQLIEQDVERYTEGEAVAEHYLAVRDA
Mvan_1167|M.vanbaalenii_PYR-1 VETFIVRSWGEHMHQHYTRLTGQDQLIEQNVEQYTDGTPVSQHYLAVRDV
MSMEG_0357|M.smegmatis_MC2_155 VEAFIVRSWEEHLRQHHTRLTGHDLLVEETVRRFTVGEPTSQHLIAAKTH
TH_1317|M.thermoresistible__bu VEAFLVRSWDEHLRQHHVRLTLVDQRVEEEVRRYAEGPPLVEHLIAVNFD
MAB_0880|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_1084|M.ulcerans_Agy99 --------------------------------------------------
Mb1288c|M.bovis_AF2122/97 -------
Rv1258c|M.tuberculosis_H37Rv -------
MMAR_4182|M.marinum_M -------
MAV_1406|M.avium_104 -------
Mflv_5163|M.gilvum_PYR-GCK R------
Mvan_1167|M.vanbaalenii_PYR-1 -------
MSMEG_0357|M.smegmatis_MC2_155 -------
TH_1317|M.thermoresistible__bu EITRRGV
MAB_0880|M.abscessus_ATCC_1997 -------
MUL_1084|M.ulcerans_Agy99 -------