For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SLQGKVALVTGAGRGIGRGIALRLAQDGADIALVDVTADGIDAVADEVAAIGSKATTFVADVSDRDAVFA AVEHAQSALGGFDIMVNNAGVALVGPVAEVTPDDAARVWGVNVNGVLWGIQAAAAKFVELGVPGRIINAS SIAGHDGFAMLGVYSASKFAVRALTQAAAKEYASSGITVNAYCPGVVGTDMWVEIDQRFADLTGAEVGAT FDQFVGGIALGRAETPEDVAAFVSYLAGPDAAYMTGQAPIIDGGLVYR*
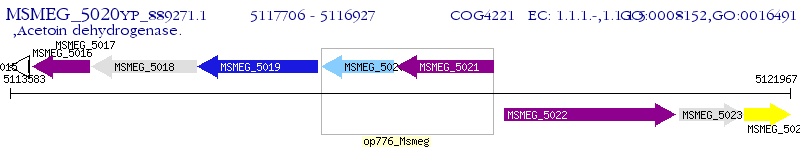
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5020 | - | - | 100% (259) | acetoin(diacetyl) reductase |
| M. smegmatis MC2 155 | MSMEG_4351 | - | 2e-39 | 40.78% (255) | oxidoreductase YjgI |
| M. smegmatis MC2 155 | MSMEG_3112 | - | 1e-35 | 36.29% (248) | estradiol 17-beta-dehydrogenase 8 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1963c | - | 2e-35 | 36.86% (255) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0124 | - | 1e-114 | 77.27% (264) | acetoin reductase |
| M. tuberculosis H37Rv | Rv1928c | - | 2e-35 | 36.86% (255) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 4e-27 | 36.07% (183) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0827 | - | 1e-38 | 40.15% (259) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_2981 | fabG3_1 | 3e-35 | 38.52% (257) | 20-beta-hydroxysteroid dehydrogenase FabG3_1 |
| M. avium 104 | MAV_1395 | - | 3e-36 | 39.44% (251) | 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2435 | - | 1e-33 | 37.90% (248) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_2885 | - | 2e-34 | 37.11% (256) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_0789 | - | 1e-114 | 76.89% (264) | acetoin reductases |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5020|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0827|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_1395|M.avium_104 --------------------------------------------------
TH_2435|M.thermoresistible__bu --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5020|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0827|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_1395|M.avium_104 --------------------------------------------------
TH_2435|M.thermoresistible__bu --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5020|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0827|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_1395|M.avium_104 --------------------------------------------------
TH_2435|M.thermoresistible__bu --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5020|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0827|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_1395|M.avium_104 --------------------------------------------------
TH_2435|M.thermoresistible__bu --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5020|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0827|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_1395|M.avium_104 --------------------------------------------------
TH_2435|M.thermoresistible__bu --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5020|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0827|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_1395|M.avium_104 --------------------------------------------------
TH_2435|M.thermoresistible__bu --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mb1963c|M.bovis_AF2122/97 -----------------MSVLDLFDLHGKRALITGASTGIGKRVALAYVE
Rv1928c|M.tuberculosis_H37Rv -----------------MSVLDLFDLHGKRALITGASTGIGKRVALAYVE
Mflv_0124|M.gilvum_PYR-GCK -----------------MA------INGKVALVTGAARGIGRGIALRLAR
Mvan_0789|M.vanbaalenii_PYR-1 -----------------MT------LNGKVALVTGAARGIGRGIALRLAR
MSMEG_5020|M.smegmatis_MC2_155 -----------------MS------LQGKVALVTGAGRGIGRGIALRLAQ
MAB_0827|M.abscessus_ATCC_1997 -----------------MTSP----LKSRRAFVTGGSRGIGAGIVRRLAE
MMAR_2981|M.marinum_M -----------------MAGR----LTGKVALVSGGARGMGASHVRALVA
MAV_1395|M.avium_104 -----------------------------MAVVTGGGSGIGRAIVERLAH
TH_2435|M.thermoresistible__bu --------------------------------VTGGASGLGRAIAARLGD
MUL_2885|M.ulcerans_Agy99 -----------------MRYP---DLAGKATIVTGAGAGIGLAIAERLAA
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
::*.. *:* .
Mb1963c|M.bovis_AF2122/97 AGAQVAIA-ARHLDALEKLADEIGTSGGKVVPVCCDVSQHQQVTSMLDQV
Rv1928c|M.tuberculosis_H37Rv AGAQVAIA-ARHLDALEKLADEIGTSGGKVVPVCCDVSQHQQVTSMLDQV
Mflv_0124|M.gilvum_PYR-GCK DGADVALV-DVHQDGLDDVAAEIAEIGSKTTTFVADVSDRDQVFAAVRHA
Mvan_0789|M.vanbaalenii_PYR-1 DGADIALV-DVLPEGLDGVADKIAGIGGKTTMFVADVSDREQVFAAVDHA
MSMEG_5020|M.smegmatis_MC2_155 DGADIALV-DVTADGIDAVADEVAAIGSKATTFVADVSDRDAVFAAVEHA
MAB_0827|M.abscessus_ATCC_1997 DGARVVFTYSSSVQQAADVVDDINEDGGWARAVKADVADPDQLGAAIGQA
MMAR_2981|M.marinum_M EGAHVVLG-DILDDEGRAVAAELGDA---ARYVHLDVTQPEQWTAAVDTA
MAV_1395|M.avium_104 DGHRVAVL-DVNEEAAEKVAARVAADGAHAIAVPTDVAESASVAAAFESV
TH_2435|M.thermoresistible__bu DGHRVAVL-DIDHAAAEQTAAELRAHGGDAVAVGADVADQDAVHDAFAAI
MUL_2885|M.ulcerans_Agy99 EGAAVLCA-DIDGASAEQAAAKIGSG---AVGYQVDVSAEEQVAGMVEAC
MLBr_00862|M.leprae_Br4923 RGAEVVVS-DIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQV
* : . : . **:
Mb1963c|M.bovis_AF2122/97 TAELGGIDIAVCNAGIITVTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAK
Rv1928c|M.tuberculosis_H37Rv TAELGGIDIAVCNAGIITVTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAK
Mflv_0124|M.gilvum_PYR-GCK SSALGGFDIMVNNAGIALVGPISEVTPAEIQKLWSINVDGVLWGIQAAVA
Mvan_0789|M.vanbaalenii_PYR-1 AAALGGFDIMVNNAGIALVGPISDVTPEQIEKLWAINVNGVLWGTQAAVA
MSMEG_5020|M.smegmatis_MC2_155 QSALGGFDIMVNNAGVALVGPVAEVTPDDAARVWGVNVNGVLWGIQAAAA
MAB_0827|M.abscessus_ATCC_1997 AEELGGLDILVNNAGILSIGDVDTATLADFDEIVAVNVRAVFAGIKAASP
MMAR_2981|M.marinum_M VNEFGGLHVLVNNAGILNIGTIEDYALSEWQRILDINVTGVFLGIRAAVK
MAV_1395|M.avium_104 RRALGSVQVLVTSAAITGFKPFAEITIEDWNRHLAVNLTGTFLCLQAALP
TH_2435|M.thermoresistible__bu RGSLGPVHILVTSAAIAGFTRFDEITLDEWNRYLAVNLTGTFLCVQAALT
MUL_2885|M.ulcerans_Agy99 VTSFGGLDKLVANAGVVHLAPILDTAVEDFDRVIGINLRGAWLCTKHAAP
MLBr_00862|M.leprae_Br4923 SAKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFGQ
* . * .*.: . . : . *: .. :
Mb1963c|M.bovis_AF2122/97 AMVKQ-----GQGGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAM
Rv1928c|M.tuberculosis_H37Rv AMVKQ-----GQGGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAM
Mflv_0124|M.gilvum_PYR-GCK QFTTLHNKEQGKISKIINASSIAG-HDG-FAMLGPYSATKFAVRALTQAA
Mvan_0789|M.vanbaalenii_PYR-1 KFKELKNKDKGRVSRIINASSIAG-HDG-FAMLGPYSATKFAVRALTQAA
MSMEG_5020|M.smegmatis_MC2_155 KFVEL-----GVPGRIINASSIAG-HDG-FAMLGVYSASKFAVRALTQAA
MAB_0827|M.abscessus_ATCC_1997 HLTEG-----GR---IVTIGSING-DSSPTANLSLYSMTKAAVAGLTRGL
MMAR_2981|M.marinum_M PMKEA-----GRGS-IINISSIEG-LAGTIASHG-YTVSKFAVRGLTKST
MAV_1395|M.avium_104 DMVEAG------WGRVVTISSTAA--QTGSPRQGHYSASKGGVIALTRTI
TH_2435|M.thermoresistible__bu DMVADG------WGRIVTISSAAG--QQGAHRQGHYAATKGGVIALTKTV
MUL_2885|M.ulcerans_Agy99 KMVERG------GGAIVNMSSLAG--HIGVGGTAAYGMSKAGISHLSRVT
MLBr_00862|M.leprae_Br4923 RLVERG-----TGGHIVNVSSMAA--YAPLQSLSAYCTSKAATYMFSDCL
: ::. .* . . * :* . ::
Mb1963c|M.bovis_AF2122/97 AVELAPHKIRVNSVSPGYILTELVEP------------YTEYQPLWEPKI
Rv1928c|M.tuberculosis_H37Rv AVELAPHKIRVNSVSPGYILTELVEP------------YTEYQPLWEPKI
Mflv_0124|M.gilvum_PYR-GCK AKEHAADGITVNAYCPGVVGTDMWVEIDKRFAELTGAAEGETYDKFVGGI
Mvan_0789|M.vanbaalenii_PYR-1 AKEHAADGITVNAYCPGVVGTDMWVEIDKRFAELTGAAEGETYDKFVGGI
MSMEG_5020|M.smegmatis_MC2_155 AKEYASSGITVNAYCPGVVGTDMWVEIDQRFADLTGAEVGATFDQFVGGI
MAB_0827|M.abscessus_ATCC_1997 AREFGPRGITINNVQPGPIGTDMNPE------------DGQLADILRPNI
MMAR_2981|M.marinum_M ALELGPSGIRVNSIHPGLVKTPMT--------------EWVPEDLFQT--
MAV_1395|M.avium_104 ALEYAAHGITANTVPPFSVDTPMLRAAQEA-------GNLPPVKYLAKAS
TH_2435|M.thermoresistible__bu ALDYAAKGITANTVPPFVIDSPMLREQQRR-------GTLPPAEYLTRAI
MUL_2885|M.ulcerans_Agy99 AAELRSANVRSNALLPAFVDTPMQQSAMAMFD----EVLGTGGANTMIAR
MLBr_00862|M.leprae_Br4923 RAELDAVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMF
: . : . * : : :
Mb1963c|M.bovis_AF2122/97 PLGRLGRPEELAGLYLYLASEASSYMTGSDIVIDGGYTCP----------
Rv1928c|M.tuberculosis_H37Rv PLGRLGRPEELAGLYLYLASEASSYMTGSDIVIDGGYTCP----------
Mflv_0124|M.gilvum_PYR-GCK ALGRAETPDDVAGFVSYLAGPDSDYMTGQAGLIDGGLVYR----------
Mvan_0789|M.vanbaalenii_PYR-1 ALGRAETPDDVAGFVSYLAGPDADYMTGQSGLIDGGLVYR----------
MSMEG_5020|M.smegmatis_MC2_155 ALGRAETPEDVAAFVSYLAGPDAAYMTGQAPIIDGGLVYR----------
MAB_0827|M.abscessus_ATCC_1997 AVGRYGQPRDIASLVSYLVSDEAWYITGTAINIDGG---------FTA--
MMAR_2981|M.marinum_M ALGRAAEPMEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHKDFSAVD
MAV_1395|M.avium_104 PVGRLGTGEDIAAACAFLCSDEAGYITGQIIGVNGGEVI-----------
TH_2435|M.thermoresistible__bu PMGRLGVAEDVAAVCSFLCSDAAGYLTGQVIGVNGGAIR-----------
MUL_2885|M.ulcerans_Agy99 LQGRMARPEEMARIVAFLLPDDSSMITGTTQIADGGTIAALW--------
MLBr_00862|M.leprae_Br4923 RLRRYGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTAR
* : . .. *
Mb1963c|M.bovis_AF2122/97 ---------
Rv1928c|M.tuberculosis_H37Rv ---------
Mflv_0124|M.gilvum_PYR-GCK ---------
Mvan_0789|M.vanbaalenii_PYR-1 ---------
MSMEG_5020|M.smegmatis_MC2_155 ---------
MAB_0827|M.abscessus_ATCC_1997 ---------
MMAR_2981|M.marinum_M VAAQPEWVT
MAV_1395|M.avium_104 ---------
TH_2435|M.thermoresistible__bu ---------
MUL_2885|M.ulcerans_Agy99 ---------
MLBr_00862|M.leprae_Br4923 IRVI-----