For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HLEGKKVVVVGGSAGIGRRVAADVVDHGGSAVVVGHSKARVDETLDDLVNRGGDAWGISAELTDRDAVTD IQRVLGEQHRDATLLVNAAGFFIPKPFLEYGQLDYDAYMELNYALFFLTQTVVRGMIAGGDGGSIVNIGS MWAHQALGVTPSTGHSMQKAGLHALTHNLAIELARHKIRVNAVSPAAVRTPALQRWFPTDEIDATLGEFA PLHPLGRVGTPTDIAGAIRYLLSDESSWITGMILNVDGGFTAGRN*
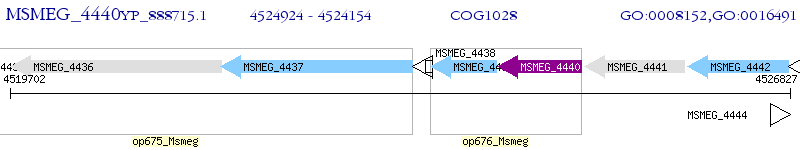
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4440 | - | - | 100% (256) | glucose-1-dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4419 | - | e-105 | 71.54% (253) | glucose-1-dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6753 | - | 4e-27 | 32.67% (251) | oxidoreductase, short chain dehydrogenase/reductase family |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1963c | - | 3e-29 | 30.56% (252) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0661 | - | 1e-25 | 30.36% (247) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1928c | - | 3e-29 | 30.56% (252) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 4e-18 | 31.08% (251) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_4680c | - | 4e-27 | 35.09% (265) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0629 | - | 3e-27 | 34.39% (253) | dehydrogenase |
| M. avium 104 | MAV_2775 | - | 5e-28 | 32.02% (253) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3505 | - | 1e-29 | 36.90% (252) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0336 | - | 7e-23 | 31.72% (268) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0489 | - | 1e-25 | 30.36% (247) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_2775|M.avium_104 --------------------------------------------------
MSMEG_4440|M.smegmatis_MC2_155 --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MMAR_0629|M.marinum_M --------------------------------------------------
Mflv_0661|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0489|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_0336|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_2775|M.avium_104 --------------------------------------------------
MSMEG_4440|M.smegmatis_MC2_155 --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MMAR_0629|M.marinum_M --------------------------------------------------
Mflv_0661|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0489|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_0336|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_2775|M.avium_104 --------------------------------------------------
MSMEG_4440|M.smegmatis_MC2_155 --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MMAR_0629|M.marinum_M --------------------------------------------------
Mflv_0661|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0489|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_0336|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
Mb1963c|M.bovis_AF2122/97 --------------------------------------------------
Rv1928c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_2775|M.avium_104 --------------------------------------------------
MSMEG_4440|M.smegmatis_MC2_155 --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MMAR_0629|M.marinum_M --------------------------------------------------
Mflv_0661|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0489|M.vanbaalenii_PYR-1 ------------------------------------------MREVRPFT
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_0336|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Mb1963c|M.bovis_AF2122/97 --MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQVAIAARHLDAL
Rv1928c|M.tuberculosis_H37Rv --MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQVAIAARHLDAL
MAV_2775|M.avium_104 ----MLDLFDLRGRRALVTGASTGIGKQVAQAYLQAGARVAIAARDFEAL
MSMEG_4440|M.smegmatis_MC2_155 --------MHLEGKKVVVVGGSAGIGRRVAADVVDHGGSAVVVGHSKARV
TH_3505|M.thermoresistible__bu -LTIAPADILLTDRVAVVTGGGAGLGRGIAAGFAAFGARVAIWERDPDSC
MMAR_0629|M.marinum_M ----------MAGLTALITGATSGIGRATAHGLAELGATVLVSGRDEARG
Mflv_0661|M.gilvum_PYR-GCK -------MTAGDRRVAIVTAAAAGIGKAIALRWCREGGCCVMADT-DGEG
Mvan_0489|M.vanbaalenii_PYR-1 NRRRVSAVKAGDRRVAIVTAAAAGIGKAIALRWCREGGCCVMADT-DGEG
MAB_4680c|M.abscessus_ATCC_199 -MNGNSALLQG--KNVIVTGGARGLGAAFSRHIVSQGGRVVIADVLDGDG
MUL_0336|M.ulcerans_Agy99 ------MVDELAGKIAIVTGGASGIGRATVARFIAEGARVVIADVEEERG
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVESAAE
.::... *:* *.
Mb1963c|M.bovis_AF2122/97 EKLADEIGTSG-GKVVPVCCDVSQHQQVTSMLDQVTAELG-GIDIAVCNA
Rv1928c|M.tuberculosis_H37Rv EKLADEIGTSG-GKVVPVCCDVSQHQQVTSMLDQVTAELG-GIDIAVCNA
MAV_2775|M.avium_104 QRTADELTAGGTGGVVAIRCDVTQPDEVSGMVDRMIAELG-GIDIAVCNA
MSMEG_4440|M.smegmatis_MC2_155 DETLDDLVNRG-GDAWGISAELTDRDAVTDIQRVLGEQHR-DATLLVNAA
TH_3505|M.thermoresistible__bu ARTAEEIG------ALGIVTDVRDSAAVSAALRQTTEQLG-SPTILVNNA
MMAR_0629|M.marinum_M REVVDEVTARG-GRGIFLAAELRDVTGVRNLATAAADAGAGKVDILINSA
Mflv_0661|M.gilvum_PYR-GCK LDAAVHELAESGAAVCGVAGDVCLSDVANSVVERALTEFG-RLDTLFNVV
Mvan_0489|M.vanbaalenii_PYR-1 LDAAVHELAESGAAVCGVAGDVCLSDVANSVVERALTEFG-RLDTLFNVV
MAB_4680c|M.abscessus_ATCC_199 RQLA----AELGDAARFTHLDVTDAEQWRELIDTTLAGFG-TITGLVNNA
MUL_0336|M.ulcerans_Agy99 ESLA----AALGADAMFCRTDVSQPEQVAAVVAAAVDNFG-GLHVMVNNA
MLBr_02565|M.leprae_Br4923 ALDETANRVGG----TTLSLDVTADDAVDKITEHLHDYQGGHADILVNNA
:: . .
Mb1963c|M.bovis_AF2122/97 GII--TVTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQGQGGVII
Rv1928c|M.tuberculosis_H37Rv GII--TVTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQGQGGVII
MAV_2775|M.avium_104 GII--SVSPMLEMPAAEFQRIQDTNVTGVFLTAQAAARAMVRQGRGGAII
MSMEG_4440|M.smegmatis_MC2_155 GFF--IPKPFLEYGQLDYDAYMELNY-ALFFLTQTVVRGMIAGGDGGSIV
TH_3505|M.thermoresistible__bu GGT--FSSPLLETSENGWDALYRANLRHVLLCTQQVGRQLVAAGQPGSIV
MMAR_0629|M.marinum_M GAF--PFGPTADTSPDDFDAVFALNVRAPYFLVGALAPTMAKRGR-GSIV
Mflv_0661|M.gilvum_PYR-GCK GGS--LARNVEEISEEQWRSQIDMNLGSVFQMSKPVIPLLRQGGG-GSIV
Mvan_0489|M.vanbaalenii_PYR-1 GGS--LARNVEEISEEQWRSQIDMNLGSVFQMSKPVIPLLRQGGG-GSIV
MAB_4680c|M.abscessus_ATCC_199 GIS--SAAMVADEPLEHFRKVIEINLVGVFIGMQAVIPAMRAHGC-GSIV
MUL_0336|M.ulcerans_Agy99 GVSGAMHRRFLDDDLADFHRVMAVNVLGVMAGTRDAARHMAAHGG-GSIV
MLBr_02565|M.leprae_Br4923 GIT--RDKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSISKG-GRVI
* : : * * ::
Mb1963c|M.bovis_AF2122/97 NTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPHKIRVNSVSPG
Rv1928c|M.tuberculosis_H37Rv NTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPHKIRVNSVSPG
MAV_2775|M.avium_104 TTASMSGHIINVPQQVGHYCASKAAVIQLTKAMAVEFAPHDIRVNSVSPG
MSMEG_4440|M.smegmatis_MC2_155 NIGSMWAHQALGVTPSTGHSMQKAGLHALTHNLAIELARHKIRVNAVSPA
TH_3505|M.thermoresistible__bu NITSIEG--ARAAPGYAAYAAAKAGVINYTRTAAFELAPHRIRVNAIAPD
MMAR_0629|M.marinum_M NVTTMVA--EFGAAGTGLYGASKAAIALLTKSWAAEYGPSGVRVNAVSPG
Mflv_0661|M.gilvum_PYR-GCK NVASTAG--ILAENRCSAYSASKGGVVLLTKNMALDFARDNIRVNAIAPG
Mvan_0489|M.vanbaalenii_PYR-1 NVASTAG--ILAENRCSAYSASKGGVVLLTKNMALDFARDNIRVNAIAPG
MAB_4680c|M.abscessus_ATCC_199 NISSAAG--LVGLALTSGYGASKWGVRGLSKVAAIELGRERIRVNSVHPG
MUL_0336|M.ulcerans_Agy99 NLTSIGG--IQAGGGVMTYRASKGAVIQFTKSAAIELAHYEIRVNAIAPG
MLBr_02565|M.leprae_Br4923 VLSSMAG--IAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPG
: . : * .: :: * . : :*:: *
Mb1963c|M.bovis_AF2122/97 YILTELVEPYTEY-------------QPLWEPKIPLGRLGRPEELAGLYL
Rv1928c|M.tuberculosis_H37Rv YILTELVEPYTEY-------------QPLWEPKIPLGRLGRPEELAGLYL
MAV_2775|M.avium_104 YIRTELVEPLHEY-------------HRQWQPKIPLGRMGRPDELAGIYL
MSMEG_4440|M.smegmatis_MC2_155 AVRTPALQRWFPTDE-------IDATLGEFAPLHPLGRVGTPTDIAGAIR
TH_3505|M.thermoresistible__bu IAVTEGLQRMSP-----------DGIPPEVGHAIPLGRPGHVDEIAGAAV
MMAR_0629|M.marinum_M PTRTEGTVEMGE-------------ALDQLAAAAPAGRPADPTEIASTIV
Mflv_0661|M.gilvum_PYR-GCK GTRTPRIERFLAEH---------PEHASMMVDLCAMQRFAGPDEIAAPAV
Mvan_0489|M.vanbaalenii_PYR-1 GTRTPRIERFLAEH---------PEHASMMVDLCAMQRFAGPDEIAAPAV
MAB_4680c|M.abscessus_ATCC_199 VIYTPMTAKEGFRE---------GEGN---MPIAAMGRVGNADEVAGAVA
MUL_0336|M.ulcerans_Agy99 NIPTPLLASSAAGLDQEQVERFTAQIRQTMREDRPLKREGTPEDIAEAAL
MLBr_02565|M.leprae_Br4923 FIETQMTAAIPLAT------------REVGRRLNSLLQGGLPVDVAETIA
* . : . ::*
Mb1963c|M.bovis_AF2122/97 YLASEASSYMTGSDIVIDGGYTCP--------------------
Rv1928c|M.tuberculosis_H37Rv YLASEASSYMTGSDIVIDGGYTCP--------------------
MAV_2775|M.avium_104 YLASAASSYMTGSDIVIDGGYSCP--------------------
MSMEG_4440|M.smegmatis_MC2_155 YLLSDESSWITGMILNVDGGFTAG-------RN-----------
TH_3505|M.thermoresistible__bu FLASEMSSYITGQTLHVDGGTQAAGGWYHHPRTGAPTLGPGPVR
MMAR_0629|M.marinum_M YLASDAASFIHGAVVPVDGGRAAV--------------------
Mflv_0661|M.gilvum_PYR-GCK FLASPEASYITGAVLPVDGGMTAGVTFRAFEAV-----------
Mvan_0489|M.vanbaalenii_PYR-1 FLASPEASYITGAVLPVDGGMTAGVTFRAFEAV-----------
MAB_4680c|M.abscessus_ATCC_199 YLLSDAASYVTGAELAVDGGWTAGPTLTTITGQ-----------
MUL_0336|M.ulcerans_Agy99 YFAGERSRYVTGTVLPVDGGTVAGKVIRRSR-------------
MLBr_02565|M.leprae_Br4923 YFAIPASNAVTGNVIRVCGQAMLGA-------------------
:: : : * : : *