For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDLGLTGKRFVVTGGTRGIGRAVVEGLLAEGAAVAFCARTPEAVTATQQELAADGVTVVGTPVDVGDGAA VADWVAQSAETLGGLDGVVANVSALAIPESPENWRTSFEVDMMGTVGLVDAALPHLLKSDAGSIVTISSV SGREIDFAAGPYGTFKAAIIHYTQGLAYQLAAKGVRANSVSPGNTYFEGGVWPTIEKDDPELFASALALN PTGRMGTPREVANAVVFLSSPVASFITGTNLVVDGALTRGVQF
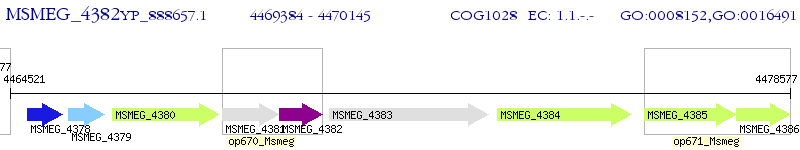
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4382 | - | - | 100% (253) | dehydrogenase/reductase SDR family protein member 10 |
| M. smegmatis MC2 155 | MSMEG_4125 | - | 8e-31 | 35.04% (254) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_1740 | - | 2e-30 | 33.06% (248) | dehydrogenase/reductase SDR family protein member 4 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0950c | - | 1e-26 | 32.93% (249) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_5148 | - | 1e-114 | 79.45% (253) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0927c | - | 1e-26 | 32.93% (249) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 3e-18 | 29.15% (247) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0833 | - | 1e-37 | 35.89% (248) | Short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4581 | - | 2e-26 | 32.51% (243) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_3019 | - | 1e-27 | 35.98% (264) | carveol dehydrogenase |
| M. thermoresistible (build 8) | TH_2589 | - | 1e-32 | 34.68% (248) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4042 | - | 8e-27 | 32.33% (266) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1185 | - | 1e-107 | 77.78% (252) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4042|M.ulcerans_Agy99 --------------MQLSFQDRTYLVTGGGSGIGKGVAAGLVAAGASVMI
MAV_3019|M.avium_104 ---------------MSGLRGRVVLVTGAGRGIGRSHCQRFAEEGADVIA
Mflv_5148|M.gilvum_PYR-GCK --------------MDLGLSGKRFVVSGGTRGIGRAVVEGLLAEGASVAF
Mvan_1185|M.vanbaalenii_PYR-1 --------------MELGLEGKRFAVTGGTRGIGRAVVEGLLAEGASVAY
MSMEG_4382|M.smegmatis_MC2_155 --------------MDLGLTGKRFVVTGGTRGIGRAVVEGLLAEGAAVAF
Mb0950c|M.bovis_AF2122/97 -----------MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLI
Rv0927c|M.tuberculosis_H37Rv -----------MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLI
MMAR_4581|M.marinum_M -----------MILDRFRLDDQVAVITGGGRGLGAAIALAFAEAGADVVI
MAB_0833|M.abscessus_ATCC_1997 -------MNRETFSRLFDLTGRTAIVTGGTRGIGRALAEGYACAGANVVV
TH_2589|M.thermoresistible__bu -------VDRQTFDKLFDMTGRTVVVTGGTRGIGLALAEGYVLAGARVVV
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
: .: ::*. *:* . * *
MUL_4042|M.ulcerans_Agy99 VGRNPDKLAGAVQELEALGANGGAIRYEPTDITNEDETARAVDAVTAWHG
MAV_3019|M.avium_104 VD-VPAAAPDLAQTAAAVQQRGVRAATALADVSDFAALAAAVDEAVGRLG
Mflv_5148|M.gilvum_PYR-GCK CARTPAAVEQAQAELSAN---GAAVAGAVVDVGDAAAVEAWVTQAAGMFS
Mvan_1185|M.vanbaalenii_PYR-1 CARTYEPVP--------------GAVGTAVDVGDGAALATWVDATADAFS
MSMEG_4382|M.smegmatis_MC2_155 CARTPEAVTATQQELAAD---GVTVVGTPVDVGDGAAVADWVAQSAETLG
Mb0950c|M.bovis_AF2122/97 ASRTSSELDAVAEQIRAA---GRRAHTVAADLAHPEVTAQLAGQAVGAFG
Rv0927c|M.tuberculosis_H37Rv ASRTSSELDAVAEQIRAA---GRRAHTVAADLAHPEVTAQLAGQAVGAFG
MMAR_4581|M.marinum_M ASRTQSQLDEVADTVRSI---GRRAHVVAADLAHPEATAELAGQAVEAFG
MAB_0833|M.abscessus_ATCC_1997 ASRKPEACTEAVERLRSL---GARALGVPTHTGDIHSLEELVRATAEEFG
TH_2589|M.thermoresistible__bu ASRKSDACERAAAHLRAL---GGDAIGVPTHLGDIDACEALVRRTVDEFG
MLBr_01807|M.leprae_Br4923 THRASVVPD--------------WFFGVECDVTDNDAIGAAFKAVEEHQG
. . .
MUL_4042|M.ulcerans_Agy99 RLHGVVHCAGGSENIGPITQVDSEAWRRTVDLNVNGTMYVLKHAAREMVR
MAV_3019|M.avium_104 RLDVLVGNAGIHPAAAPAWEITPQNWQQTLDVNLTGVWHTVKAGVPHMSR
Mflv_5148|M.gilvum_PYR-GCK GLDGVVANVS-----ALAIPETHENWRTTFEVDLMGTVGMVDAAMPYLLE
Mvan_1185|M.vanbaalenii_PYR-1 GLDGVVANVS-----ALAIPETHENWHTSFDVDLMGTVNLVDAALPYLLE
MSMEG_4382|M.smegmatis_MC2_155 GLDGVVANVS-----ALAIPESPENWRTSFEVDMMGTVGLVDAALPHLLK
Mb0950c|M.bovis_AF2122/97 KLDIVVNNVGGT-MPNTLLSTSTKDLADAFAFNVGTAHALTVAAVPLMLE
Rv0927c|M.tuberculosis_H37Rv KLDIVVNNVGGT-MPNTLLSTSTKDLADAFAFNVGTAHALTVAAVPLMLE
MMAR_4581|M.marinum_M RLDIVVNNVGGT-MPNTLLTTSTKDLKDAFTFNVATAHALTVAAVPLMLE
MAB_0833|M.abscessus_ATCC_1997 GIDIVVNNAANA-LTEPVGAFTVDGWDKSFGVNLRGPVFLVQAALPHLVA
TH_2589|M.thermoresistible__bu GIDVLVNNAANA-LTQPLGELTVDAWTKSFEVNLRGPVFLVQAALPHLKT
MLBr_01807|M.leprae_Br4923 PVEVLVANAGIT-SDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIK
:. :* .. . . .:: . :
MUL_4042|M.ulcerans_Agy99 GG-GGSFVGISSIAASNTHRWFGAYGVTKSAVDHLMQLAADELGASWVRV
MAV_3019|M.avium_104 TARGGSIVIISSTSGIRGTPGAAPYSASKHAVVGLARTLANELGPQGIRV
Mflv_5148|M.gilvum_PYR-GCK SD-AGSIVTVSSVSGREVDFAAGPYGTMKAAIIHYTQGLAFKYAGQGVRA
Mvan_1185|M.vanbaalenii_PYR-1 AD-SGSIVTISSVSGREIDFAAGPYGTMKAAIIHYTQGLAFKLAAQGVRA
MSMEG_4382|M.smegmatis_MC2_155 SD-AGSIVTISSVSGREIDFAAGPYGTFKAAIIHYTQGLAYQLAAKGVRA
Mb0950c|M.bovis_AF2122/97 HSGGGSVINISSTMGRLAARGFAAYGTAKAALAHYTRLAALDLCPR-VRV
Rv0927c|M.tuberculosis_H37Rv HSGGGSVINISSTMGRLAARGFAAYGTAKAALAHYTRLAALDLCPR-VRV
MMAR_4581|M.marinum_M HSGGGSIINITSTMGRVAGRGFAAYGTAKAALAHYTRLAALDLCPR-IRV
MAB_0833|M.abscessus_ATCC_1997 SP-HAAVLNVVSVGAFMFGQGVSMYSAAKAALVSYTRSAAAELASRGVRV
TH_2589|M.thermoresistible__bu SP-HAAVLNMVSVGAFLFSPGTAMYSAGKAALMSFTRSMAAQFAPLGIRV
MLBr_01807|M.leprae_Br4923 QR-FGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITA
. : : * . . *.. * .: : * . : .
MUL_4042|M.ulcerans_Agy99 NSIRPGLIRTD---LVAAITESAELSSDYAMCTPLPRQG-----------
MAV_3019|M.avium_104 NTVHPGAVATA---MVLNEATFRRLRPDLDNPTADDAAEALSARHLLPVP
Mflv_5148|M.gilvum_PYR-GCK NTVSPGNTYFPGGVWTSIEENDPQLFATALALNPTGRMA-----------
Mvan_1185|M.vanbaalenii_PYR-1 NTVSPGNTYFPGGVWPSIEQSDPELFATALALNPTGRMA-----------
MSMEG_4382|M.smegmatis_MC2_155 NSVSPGNTYFEGGVWPTIEKDDPELFASALALNPTGRMG-----------
Mb0950c|M.bovis_AF2122/97 NAIAPGSILTS---ALEVVAANDELRAPMEQATPLRRLG-----------
Rv0927c|M.tuberculosis_H37Rv NAIAPGSILTS---ALEVVAANDELRAPMEQATPLRRLG-----------
MMAR_4581|M.marinum_M NAIAPGSILTS---ALEVVAANDELRAPMEQATPLRRLG-----------
MAB_0833|M.abscessus_ATCC_1997 NALAPGAIDTD---MVRKTPP--AEVERMANTNHLKRLG-----------
TH_2589|M.thermoresistible__bu NALAPGPVDTD---MMRNNPQ--EVVDMMAGATLMKRLA-----------
MLBr_01807|M.leprae_Br4923 NVVAPGFIDTE-----MTRAMDSKYQEAALQAIPLGRIG-----------
* : **
MUL_4042|M.ulcerans_Agy99 --EVEDVANMAMFLLSDAASFVTGQVINVDGGQMLRRGPDFSAMLEPVFG
MAV_3019|M.avium_104 WVEPVDVSNAVVFLASDQARYITGTQIVVDAGLLAKA-------------
Mflv_5148|M.gilvum_PYR-GCK --TPQEVANAVVFLSSPAASFITGTNLLVDGALTRGVQF-----------
Mvan_1185|M.vanbaalenii_PYR-1 --TPQEVANAVVFLSSAAASFITGTNLLVDGALTRGVQL-----------
MSMEG_4382|M.smegmatis_MC2_155 --TPREVANAVVFLSSPVASFITGTNLVVDGALTRGVQF-----------
Mb0950c|M.bovis_AF2122/97 --DPVDIAAAAVYLASPAGSFLTGKTLEVDGGLTFPNLDLPIPDL-----
Rv0927c|M.tuberculosis_H37Rv --DPVDIAAAAVYLASPAGSFLTGKTLEVDGGLTFPNLDLPIPDL-----
MMAR_4581|M.marinum_M --DPVDIAAAAVYLASPAGSFLTGKTLEVDGGLTHPNLDIPVPDL-----
MAB_0833|M.abscessus_ATCC_1997 --LPDEMVGTALLLTSDAGSYITGQTIIADGGLVAR--------------
TH_2589|M.thermoresistible__bu --TPDEMVGAALLLTSDAGSYITGTVLIADGGGTPR--------------
MLBr_01807|M.leprae_Br4923 --AVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH-------------
:: . * * . :::* : .*..
MUL_4042|M.ulcerans_Agy99 RDALRGLV
MAV_3019|M.avium_104 --------
Mflv_5148|M.gilvum_PYR-GCK --------
Mvan_1185|M.vanbaalenii_PYR-1 --------
MSMEG_4382|M.smegmatis_MC2_155 --------
Mb0950c|M.bovis_AF2122/97 --------
Rv0927c|M.tuberculosis_H37Rv --------
MMAR_4581|M.marinum_M --------
MAB_0833|M.abscessus_ATCC_1997 --------
TH_2589|M.thermoresistible__bu --------
MLBr_01807|M.leprae_Br4923 --------