For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHQTIQPRTTSPTRRTPQLIGTALGHALEWYDWGIYAIFVPFFATQFFDDSNQLSAVLSSLAVFAVGFVA RPVGGFVFGWLADRAGRRTAMMATVVTIAAASFAIGVAPTYAEIGAWASLILLLARIVQGLACGGELPSA QTYLSEIAPANRRGVWSSIIYISSVFGNTVGVLLGLVLTLTLSEADMHAYGWRIPFLLGGVFGLVAIYMR RKMPETEAFIQADRRRPRLWPELVRHRGQALRVIGLSVGFTVFYYSWVVAAPAYAISSLHVSSAGALWAG VTSSILLMALMPLWGRLSDRIGRKPVLLISTVGGAATLFPVQFLVRDSAWQLFAGMTIAAVFISAGVSIL PAVYAEMFPTGIRALGLAVPYSVAVAVFGGTAPYLQTWIGANLGPLFYTGYCVLLLLASTLVIIRMPETR GKDLT
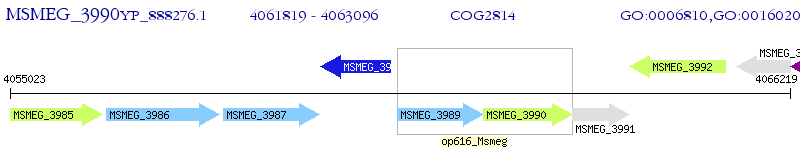
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3990 | - | - | 100% (425) | putative transporter |
| M. smegmatis MC2 155 | MSMEG_2952 | - | e-108 | 44.79% (413) | transport gene |
| M. smegmatis MC2 155 | MSMEG_2486 | - | 4e-68 | 33.99% (406) | major facilitator superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3503c | kgtP | 2e-61 | 35.65% (418) | dicarboxylic acid transport integral membrane protein KgtP |
| M. gilvum PYR-GCK | Mflv_0953 | - | 3e-71 | 34.38% (413) | general substrate transporter |
| M. tuberculosis H37Rv | Rv3476c | kgtP | 8e-61 | 35.41% (418) | dicarboxylic acid transport integral membrane protein KgtP |
| M. leprae Br4923 | MLBr_01562 | - | 2e-06 | 28.89% (135) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_3808c | - | 8e-65 | 35.39% (421) | dicarboxylic acid transport integral membrane protein KgtP |
| M. marinum M | MMAR_4238 | - | 1e-39 | 29.79% (423) | integral membrane transport protein |
| M. avium 104 | MAV_4627 | - | 8e-49 | 32.67% (401) | major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_3400 | - | 3e-75 | 35.44% (412) | PUTATIVE General substrate transporter |
| M. ulcerans Agy99 | MUL_0949 | - | 4e-40 | 29.79% (423) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_5839 | - | 8e-70 | 33.74% (406) | general substrate transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3503c|M.bovis_AF2122/97 ----------MTVSIAPPSRPS-QAETRRAIWNTIRGSSGNLVEWYDVYV
Rv3476c|M.tuberculosis_H37Rv ----------MTVSIAPPSRPS-QAETRRAIWNTIRGSSGNLVEWYDVYV
MAB_3808c|M.abscessus_ATCC_199 ----------MAVSAADTSTPSGRAQTRRAIWNTIRGSSGNLVEWYDVYV
MSMEG_3990|M.smegmatis_MC2_155 ----------MHQTIQPRTTSP-----TRRTPQLIGTALGHALEWYDWGI
MMAR_4238|M.marinum_M -----------------------------MSRVALACVVGSTVEYYDFCI
MUL_0949|M.ulcerans_Agy99 -----------------------------MSRVALACVVGSTVEYYDFCI
MAV_4627|M.avium_104 MAATLPAPAPAGLADGHPLDESAEQQRRRLRIVVAASLLGTTVEWYDFFL
Mflv_0953|M.gilvum_PYR-GCK ----------MDSRDARPPTIEKEQPPAVLKKAIAASAIGNATEWFDYGI
Mvan_5839|M.vanbaalenii_PYR-1 ---------------MGMPTTHEDPPPGVLKKAISASAIGNATEWFDYGI
TH_3400|M.thermoresistible__bu -------LTTERTDPPQDPSVEQEQPPGVLRKAIAASAVGNATEWFDYGL
MLBr_01562|M.leprae_Br4923 ----MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLP
::
Mb3503c|M.bovis_AF2122/97 YTVFAT--YFEDQFFDRA-DRNSTVY-VYAIFAVTFVTRPVGS-------
Rv3476c|M.tuberculosis_H37Rv YTVFAT--YFEDQFFDRA-DRNSTVY-VYAIFAVTFVTRPVGS-------
MAB_3808c|M.abscessus_ATCC_199 YTVFAM--YFEKQFFDDS-DQNSTVY-VYAIFAVTFITRPLGS-------
MSMEG_3990|M.smegmatis_MC2_155 YAIFVP--FFATQFFDDS-NQLSAVLSSLAVFAVGFVARPVGG-------
MMAR_4238|M.marinum_M YGTAAA--LVFPAVFYPGLSPALATIASMGTFATAFLSRPLGA-------
MUL_0949|M.ulcerans_Agy99 YGTAAA--LVFPAVFYPGLSPALATIASMGTFATAFLSRPLGA-------
MAV_4627|M.avium_104 YATAAG--LVFNKVFFPNESSLVGTLLAFATFAVGFVMRPIGG-------
Mflv_0953|M.gilvum_PYR-GCK YAYGVT--YISAAIFPGD--AANATLLALMTFAVSFLVRPLGG-------
Mvan_5839|M.vanbaalenii_PYR-1 YAYGVS--YISAAIFPGD--AANATLLALMTFAVSFLVRPLGG-------
TH_3400|M.thermoresistible__bu YAYGVT--YISVAIFPGD--GATATLLALMTFAVSFLVRPLGG-------
MLBr_01562|M.leprae_Br4923 SGRFPSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSW
. .. .* : . :.
Mb3503c|M.bovis_AF2122/97 -------------WFFGRFADRRGRRAALTFSVSLMAACSLIVALVPSRS
Rv3476c|M.tuberculosis_H37Rv -------------WFLGRFADRRGRRAALTFSVSLMAACSLIVALVPSRS
MAB_3808c|M.abscessus_ATCC_199 -------------WFFGRFADRNGRRAALTLSVSLMAACSMVIALVPSRG
MSMEG_3990|M.smegmatis_MC2_155 -------------FVFGWLADRAGRRTAMMATVVTIAAASFAIGVAPTYA
MMAR_4238|M.marinum_M -------------AVFGHFGDRLGRKKTLVATLLIMALSTVTVGVVPSTA
MUL_0949|M.ulcerans_Agy99 -------------AVFGHFGDRLGRKKTLVATLLIMALSTVTVGVVPSTA
MAV_4627|M.avium_104 -------------LVFGHIGDRIGRKRSLALTMLIMGGATALIGVLPTAA
Mflv_0953|M.gilvum_PYR-GCK -------------FVWGPLGDRLGRKRVLAMTILLMAGATFCVGLVPTYA
Mvan_5839|M.vanbaalenii_PYR-1 -------------FVWGPLGDRLGRKRVLAITIVLMAGATFCVALVPTYD
TH_3400|M.thermoresistible__bu -------------FVWGPLGDRLGRRQVLAITILLMAGATFCVGLVPSYD
MLBr_01562|M.leprae_Br4923 GITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVA----
. * :.* **: : : : .: .:
Mb3503c|M.bovis_AF2122/97 SIGVAAPILLILCRLVQGFATGGEYGT-----SVTYMSEAATRERRGYFS
Rv3476c|M.tuberculosis_H37Rv SIGVAAPILLILCRLVQGFATGGEYGT-----SATYMSEAATRERRGYFS
MAB_3808c|M.abscessus_ATCC_199 SIGMAAPIVLILCRLVQGFATGGEYGT-----SATYMSEAATRERRGFFS
MSMEG_3990|M.smegmatis_MC2_155 EIGAWASLILLLARIVQGLACGGELPS-----AQTYLSEIAPANRRGVWS
MMAR_4238|M.marinum_M TIGVSAPLILVALRLLQGFAVGGEWAG-----SALLSIESAPAHKRGYYG
MUL_0949|M.ulcerans_Agy99 TIGVSAPLILVALRLLQGFAVGGEWAG-----SALLSIESAPAHKRGYYG
MAV_4627|M.avium_104 QIGAWAPVLLLVLRVLQGFALGGEWGG-----AVLLAVEHSPGDRRGRYG
Mflv_0953|M.gilvum_PYR-GCK AIGMWAPFLLVLLRMIQGFSTGGEYGG-----AATFMAEYSPSRRRGLLG
Mvan_5839|M.vanbaalenii_PYR-1 MIGMWAPFLLVLLRMIQGFSTGGEYGG-----AATFMAEYAPNRRRGLLG
TH_3400|M.thermoresistible__bu SIGLWAPVLMVILRMIQGFSTGGEYGG-----AATFMAEYAPSRRRGLLG
MLBr_01562|M.leprae_Br4923 ----WDEVTLVIARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFA
. :: *::**:. . : : . .
Mb3503c|M.bovis_AF2122/97 SFQYVTLVGGHVLA-------QFTLLVILAVFT--REQVHEFGWRIGFAV
Rv3476c|M.tuberculosis_H37Rv SFQYVTLVGGHVLA-------QFTLLVILAVFT--REQVHEFGWRIGFAV
MAB_3808c|M.abscessus_ATCC_199 SFQYVTLVGGHVLA-------QFTLLIILTVFD--TAQVHEFGWRIAFAT
MSMEG_3990|M.smegmatis_MC2_155 SIIYISSVFGNTVG-------VLLGLVLTLTLS--EADMHAYGWRIPFLL
MMAR_4238|M.marinum_M MFTVLGGGVALVVS-------GLTFLGVNYTIGESSSAFLQWGWRVPFLL
MUL_0949|M.ulcerans_Agy99 MFTVLGGGVALVVS-------GLTFLGVNYTIGESSSALLQWGWRVPFLL
MAV_4627|M.avium_104 AVPQVGLALGLALG-------TGIFAFLQIVLG--PARFLSYGWRIGFLL
Mflv_0953|M.gilvum_PYR-GCK SFLEFGTLAGFSCG-------ALLMLGCSLVLG--DEDMQAWGWRLPFLV
Mvan_5839|M.vanbaalenii_PYR-1 SFLEFGTLGGFSLG-------ALLMLGCSLVLG--DEQMHAWGWRLPFLV
TH_3400|M.thermoresistible__bu SFLEFGTLAGFSLG-------ALMMLGVSLMLS--DDQMNTWGWRVPFLI
MLBr_01562|M.leprae_Br4923 AMTAIGSVMGLVVGGALTEVSWRLAFLVNVPIGLVMIYLARTALHETHKE
. . . . : . . : .
Mb3503c|M.bovis_AF2122/97 GGGAAIVVFWLRRTMDESLSQE----------RLTAIKAGRDHDSGSLRE
Rv3476c|M.tuberculosis_H37Rv GGGAAIVVFWLRRTMDESLSQE----------RLTAIKAGRDHDSGSLRE
MAB_3808c|M.abscessus_ATCC_199 GGVAAIVVYWLRRTMDESLSEE----------QLAAIKAGADTSSGSMRE
MSMEG_3990|M.smegmatis_MC2_155 GGVFGLVAIYMRRKMPETEAFI----------QADRRRP-------RLWP
MMAR_4238|M.marinum_M SALLIAFALYVRLEINETPVFARAM-------AQADDQESTAAARAPLAA
MUL_0949|M.ulcerans_Agy99 SALLIAFALYVRLEINETPVFARAM-------AQADDQESTAAARAPLAA
MAV_4627|M.avium_104 SVLLVAIGIVVRLRVEETPAFR-------------ELQDLQAASTVPIRD
Mflv_0953|M.gilvum_PYR-GCK AAPLGLIGVYLRSRLEDTPVYR----------ELEQEGRTEESTSMQFRD
Mvan_5839|M.vanbaalenii_PYR-1 AAPLGLIGLYLRSRLEDTPVFR----------ELEAKGGTEPETTTQFRD
TH_3400|M.thermoresistible__bu AAPLGLIGLYLRSRLDDTPIFR----------ELQESDREEKSIWTEFKD
MLBr_01562|M.leprae_Br4923 RMKLDAAGAILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAF
: : :
Mb3503c|M.bovis_AF2122/97 LAT-------------HYWKPLLLCFLVTLGGTVAFYTYSVNAPAIVKSV
Rv3476c|M.tuberculosis_H37Rv LAT-------------HYWKPLLLCFLVTLGGTVAFYTYSVNAPAIVKSV
MAB_3808c|M.abscessus_ATCC_199 LLT-------------RYRKPLLLCFLITMGGTLAFYTYSVNAPAIVKAA
MSMEG_3990|M.smegmatis_MC2_155 ELV-------------RHRGQALRVIGLSVGFTVFYYSWVVAAPAYAISS
MMAR_4238|M.marinum_M VLC-------------RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLH
MUL_0949|M.ulcerans_Agy99 VLC-------------RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLH
MAV_4627|M.avium_104 ILRE-----------RRSRRNTVLGLLSRWAEGSAFNTWGVFAISYATGA
Mflv_0953|M.gilvum_PYR-GCK LVV-------------RYWRPILQMGGLVVALNVVNYTLLTYMPTYLENS
Mvan_5839|M.vanbaalenii_PYR-1 LLG-------------RYWRPILQLGGLVVALNVVNYTLLSYMPTYLENR
TH_3400|M.thermoresistible__bu LLV-------------EYRGQILRLGGLVVALNVVNYTLLSYMPTYLQTS
MLBr_01562|M.leprae_Br4923 AVVERTAENPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDI
:
Mb3503c|M.bovis_AF2122/97 YGSQAMTATWINLVGLILLMMLQPIGGMISDKIGRKPLLLWFGVGGLIYT
Rv3476c|M.tuberculosis_H37Rv YGSQAMTATWINLVGLILLMMLQPIGGMISDKIGRKPLLLWFGVGGLIYT
MAB_3808c|M.abscessus_ATCC_199 YKDQAMTATWINLAGLIFLMLLQPVGGIISDKVGRKPLLLFFGFGGVVYT
MSMEG_3990|M.smegmatis_MC2_155 LHVSSAGALWAGVTSSILLMALMPLWGRLSDRIGRKPVLLISTVGGAATL
MMAR_4238|M.marinum_M LGYSRNLVLLIGVLGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWS
MUL_0949|M.ulcerans_Agy99 LGYSRNLVLLIGVLGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWS
MAV_4627|M.avium_104 LGLNRVSVLIAVTIAALLMAVLLPVSGVLTDRFGARRVYLAGIACYGLAA
Mflv_0953|M.gilvum_PYR-GCK IGLSADQSLMVPIIGMLAMMVFVPFAGLLSDRVGRKPLWWFSLIGLFVAG
Mvan_5839|M.vanbaalenii_PYR-1 IGLSPDQSLIVPVIGMLSMMVFVPFAGLLSDRVGRKPLWWFSLIGLFVAG
TH_3400|M.thermoresistible__bu IGLSTDTALVVPIIGMLSMMVFLPFAGLLSDRFGRKPMWWISLGGLLALV
MLBr_01562|M.leprae_Br4923 LGYSALRAGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAM
. : . . : .:.. : :
Mb3503c|M.bovis_AF2122/97 YVLVTYLPETRSPTMSFLLVAVGYVILTGYCSINALVKSELFPAHVRALG
Rv3476c|M.tuberculosis_H37Rv YVLVTYLPETRSPTMSFLLVAVGYVILTGYCSINALVKSELFPAHVRALG
MAB_3808c|M.abscessus_ATCC_199 YVLITYLPQVHAPIVSFLLVAVSYVLLTGYTSINALVKSELFPSHVRALG
MSMEG_3990|M.smegmatis_MC2_155 FPVQFLVRDSAWQLFAGMTIAAVFISAG--VSILPAVYAEMFPTGIRALG
MMAR_4238|M.marinum_M PLVMPLIGSGDTVGFTVALLGLY-AVAASGFGPAASLIPELFATRYRYTG
MUL_0949|M.ulcerans_Agy99 PLVMPLIGSGDTVGFTVALLGLY-AVAASGFGPAASLIPELFATRYRYTG
MAV_4627|M.avium_104 FPAFALFGTKKLLWFGLAMVIVFGVVHALFYGAQGTLYSALYPTNTRYTG
Mflv_0953|M.gilvum_PYR-GCK IPAFLLMGTN--LAGAVIGFAILGLLYVPQLATISATFPAMFPTQVRYAG
Mvan_5839|M.vanbaalenii_PYR-1 VPMFLLMGTN--LWGAVIGFAVLGLLYVPQLATISATFPAMFPTQVRYAG
TH_3400|M.thermoresistible__bu VPMFMLMSTS--VPGAIIGFAVLGLLYVPQLATISATFPAMFPTHVRYAG
MLBr_01562|M.leprae_Br4923 LYGWAFMNRG-VPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIG
. . . : *
Mb3503c|M.bovis_AF2122/97 VGVGYALANSVFG-------------------GTAPLIYQALKERDQVPM
Rv3476c|M.tuberculosis_H37Rv VGVGYALANSVFG-------------------GTAPLIYQALKERDQVPM
MAB_3808c|M.abscessus_ATCC_199 VGVGYALANSVFG-------------------GTAPLIYQALKEHDQVPL
MSMEG_3990|M.smegmatis_MC2_155 LAVPYSVAVAVFG-------------------GTAPYLQTWIGANLGPLF
MMAR_4238|M.marinum_M TALALNVAGIVGG-------------------AAPPLIAGTLLASYGSS-
MUL_0949|M.ulcerans_Agy99 TALALNVAGIVGG-------------------AAPPLIAGTLLASYGSS-
MAV_4627|M.avium_104 LSFVYQFSGVYAS-------------------GVTPMILTALIAALGG--
Mflv_0953|M.gilvum_PYR-GCK FAIAYNVSTSIFG-------------------GTAPAVNDWLVGATGDNL
Mvan_5839|M.vanbaalenii_PYR-1 FAIAYNVSTSLFG-------------------GTAPAINQWLTGETGDLL
TH_3400|M.thermoresistible__bu FAIAYNVSTSLFG-------------------GTAPAANDWLIQQTGNNL
MLBr_01562|M.leprae_Br4923 PVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRAL
. . . .. :
Mb3503c|M.bovis_AF2122/97 FIAYVTACIAVSLIVYVFFIKNKADTYLDREQGFAFYGHA----------
Rv3476c|M.tuberculosis_H37Rv FIAYVTACIAVSLIVYVFFIKNKADTYLDREQGFAFYGHA----------
MAB_3808c|M.abscessus_ATCC_199 FIGYVTVCIAISLVVYLFFLKNKSQTYLDREQGSAFNR------------
MSMEG_3990|M.smegmatis_MC2_155 YTGYCVLLLLASTLVIIRMPETRGKDLT----------------------
MMAR_4238|M.marinum_M AVGLMMTALVAASLLCTYLLPETAGAPLTAA-------------------
MUL_0949|M.ulcerans_Agy99 AVGLMMTALVAASLLCTYLLPETAGAPLTAA-------------------
MAV_4627|M.avium_104 APWLACGYLVATAVISVLATALIRDRDLYL--------------------
Mflv_0953|M.gilvum_PYR-GCK VPAYYMMGACVIGALALIRIPETTRCPLNGTATPGTDEAPAPVEYEKSSA
Mvan_5839|M.vanbaalenii_PYR-1 FPAYYMMGACVIGAIALIKVPETARCPIGGTVTPGTEEAADPVPFEKQNA
TH_3400|M.thermoresistible__bu MPAFYMMAACLVGMLALVKLPETARCPIGGTEIPGTEDAPPPVEYDLVGN
MLBr_01562|M.leprae_Br4923 DHGYTYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL---
:
Mb3503c|M.bovis_AF2122/97 -
Rv3476c|M.tuberculosis_H37Rv -
MAB_3808c|M.abscessus_ATCC_199 -
MSMEG_3990|M.smegmatis_MC2_155 -
MMAR_4238|M.marinum_M -
MUL_0949|M.ulcerans_Agy99 -
MAV_4627|M.avium_104 -
Mflv_0953|M.gilvum_PYR-GCK -
Mvan_5839|M.vanbaalenii_PYR-1 -
TH_3400|M.thermoresistible__bu R
MLBr_01562|M.leprae_Br4923 -