For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVSIAPPSRPSQAETRRAIWNTIRGSSGNLVEWYDVYVYTVFATYFEDQFFDRADRNSTVYVYAIFAVTF VTRPVGSWFLGRFADRRGRRAALTFSVSLMAACSLIVALVPSRSSIGVAAPILLILCRLVQGFATGGEYG TSATYMSEAATRERRGYFSSFQYVTLVGGHVLAQFTLLVILAVFTREQVHEFGWRIGFAVGGGAAIVVFW LRRTMDESLSQERLTAIKAGRDHDSGSLRELATHYWKPLLLCFLVTLGGTVAFYTYSVNAPAIVKSVYGS QAMTATWINLVGLILLMMLQPIGGMISDKIGRKPLLLWFGVGGLIYTYVLVTYLPETRSPTMSFLLVAVG YVILTGYCSINALVKSELFPAHVRALGVGVGYALANSVFGGTAPLIYQALKERDQVPMFIAYVTACIAVS LIVYVFFIKNKADTYLDREQGFAFYGHA*
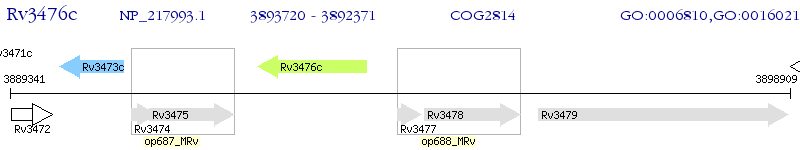
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3476c | kgtP | - | 100% (449) | PROBABLE DICARBOXYLIC ACID TRANSPORT INTEGRAL MEMBRANE PROTEIN KGTP (DICARBOXYLATE TRANSPORTER) |
| M. tuberculosis H37Rv | Rv1200 | - | 9e-31 | 28.20% (383) | integral membrane transport protein |
| M. tuberculosis H37Rv | Rv1902c | nanT | 8e-16 | 24.94% (397) | sialic acid-transport integral membrane protein NanT |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3503c | kgtP | 0.0 | 99.55% (449) | dicarboxylic acid transport integral membrane protein KgtP |
| M. gilvum PYR-GCK | Mflv_2381 | - | 1e-57 | 32.86% (420) | general substrate transporter |
| M. leprae Br4923 | MLBr_01562 | - | 6e-05 | 20.85% (307) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_3808c | - | 0.0 | 81.39% (446) | dicarboxylic acid transport integral membrane protein KgtP |
| M. marinum M | MMAR_4238 | - | 1e-31 | 25.93% (405) | integral membrane transport protein |
| M. avium 104 | MAV_1344 | - | 2e-34 | 29.79% (376) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_0985 | - | 0.0 | 80.87% (439) | sugar transporter family protein |
| M. thermoresistible (build 8) | TH_3400 | - | 2e-55 | 30.28% (426) | PUTATIVE General substrate transporter |
| M. ulcerans Agy99 | MUL_0949 | - | 5e-32 | 25.93% (405) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_4266 | - | 2e-58 | 32.63% (429) | general substrate transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3476c|M.tuberculosis_H37Rv ---------------MTVS--IAPPSRPSQAETRRAIWNTIRGSS-----
Mb3503c|M.bovis_AF2122/97 ---------------MTVS--IAPPSRPSQAETRRAIWNTIRGSS-----
MAB_3808c|M.abscessus_ATCC_199 ---------------MAVS-AADTSTPSGRAQTRRAIWNTIRGSS-----
MSMEG_0985|M.smegmatis_MC2_155 ---------------MTVAGQAHPPTRSGAAETRRAIWNTIRGSS-----
MMAR_4238|M.marinum_M -----------------------------------MSRVALACVV-----
MUL_0949|M.ulcerans_Agy99 -----------------------------------MSRVALACVV-----
MAV_1344|M.avium_104 ---------------------MGEPSARATTPGTPMRRIAAACLV-----
Mflv_2381|M.gilvum_PYR-GCK -------------MTVDEDQRRPS-----GAEPAAVRQAVRGAAI-----
Mvan_4266|M.vanbaalenii_PYR-1 -------------MTVKET--RP-------AAPGAVSRAVRGAAI-----
TH_3400|M.thermoresistible__bu -------------LTTERTDPPQDPSVEQEQPPGVLRKAIAASAV-----
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Rv3476c|M.tuberculosis_H37Rv ------GNLVEWYDVYVYTVFATYFEDQFFDR--ADRNSTVYVYAIFAVT
Mb3503c|M.bovis_AF2122/97 ------GNLVEWYDVYVYTVFATYFEDQFFDR--ADRNSTVYVYAIFAVT
MAB_3808c|M.abscessus_ATCC_199 ------GNLVEWYDVYVYTVFAMYFEKQFFDD--SDQNSTVYVYAIFAVT
MSMEG_0985|M.smegmatis_MC2_155 ------GNLVEWYDVYVYTVFATYFEKQFFAE--GDENSTVYVYAIFAVT
MMAR_4238|M.marinum_M ------GSTVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATA
MUL_0949|M.ulcerans_Agy99 ------GSTVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATA
MAV_1344|M.avium_104 ------GSAIEFYDFLIYGTAAALVFPAVFFPRLGPTVATIASMATFATA
Mflv_2381|M.gilvum_PYR-GCK ------GNTVEWFDFAIYGFLATYIA-DKFFPSGDETAALLSTFAVFAAA
Mvan_4266|M.vanbaalenii_PYR-1 ------GNAVEWFDFAIYGFLATYIA-EKFFPPGDETAALLNTFAIFAAA
TH_3400|M.thermoresistible__bu ------GNATEWFDYGLYAYGVTYIS-VAIFP-GDGATATLLALMTFAVS
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
. . : . : .:
Rv3476c|M.tuberculosis_H37Rv FVTRPVGSWFLG-RFADRRGRRAALTFSVSLMAAC---------SLIVAL
Mb3503c|M.bovis_AF2122/97 FVTRPVGSWFFG-RFADRRGRRAALTFSVSLMAAC---------SLIVAL
MAB_3808c|M.abscessus_ATCC_199 FITRPLGSWFFG-RFADRNGRRAALTLSVSLMAAC---------SMVIAL
MSMEG_0985|M.smegmatis_MC2_155 FLTRPIGSWFFG-RFADRRGRRAALTFSVSLMALC---------SLVIAI
MMAR_4238|M.marinum_M FLSRPLGAAVFG-HFGDRLGRKKTLVATLLIMALS---------TVTVGV
MUL_0949|M.ulcerans_Agy99 FLSRPLGAAVFG-HFGDRLGRKKTLVATLLIMALS---------TVTVGV
MAV_1344|M.avium_104 FLSRPLGAAVFG-YFGDRLGRKKTLVATLLIMGAS---------TVSVGL
Mflv_2381|M.gilvum_PYR-GCK FFMRPLGGFFFG-PLGDRIGRQRVLAIVILLMSAS---------TLAIGL
Mvan_4266|M.vanbaalenii_PYR-1 FFMRPLGGFFFG-PLGDRIGRQRVLALVILLMSAS---------TLAIGL
TH_3400|M.thermoresistible__bu FLVRPLGGFVWG-PLGDRLGRRQVLAITILLMAGA---------TFCVGL
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
:. * . * :.* **: .: : :: . . : :
Rv3476c|M.tuberculosis_H37Rv VPSRSSIGVAAPILLILCRLVQGFATG-GEYGTSATYMSEAATRERRGYF
Mb3503c|M.bovis_AF2122/97 VPSRSSIGVAAPILLILCRLVQGFATG-GEYGTSVTYMSEAATRERRGYF
MAB_3808c|M.abscessus_ATCC_199 VPSRGSIGMAAPIVLILCRLVQGFATG-GEYGTSATYMSEAATRERRGFF
MSMEG_0985|M.smegmatis_MC2_155 VPSREVIGAAAPVILILCRLVQGFATG-GEYGTSATYMSEAATRERRGFF
MMAR_4238|M.marinum_M VPSTATIGVSAPLILVALRLLQGFAVG-GEWAGSALLSIESAPAHKRGYY
MUL_0949|M.ulcerans_Agy99 VPSTATIGVSAPLILVALRLLQGFAVG-GEWAGSALLSIESAPAHKRGYY
MAV_1344|M.avium_104 VPSTASIGIAAPLLLTVLRLLQGFAVG-AEWAGSVLLSAEYAPTDRRGWY
Mflv_2381|M.gilvum_PYR-GCK VPSYDSIGVLAPILLLVLRCLQGFSAG-GEYGSGACFLAEYAPNRHRGFV
Mvan_4266|M.vanbaalenii_PYR-1 VPGYDSIGVLAPILLLLLRCLQGFSAG-GEYGSGACFLAEYAPDRHRGFV
TH_3400|M.thermoresistible__bu VPSYDSIGLWAPVLMVILRMIQGFSTG-GEYGGAATFMAEYAPSRRRGLL
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
. ** . *: * .. . * *
Rv3476c|M.tuberculosis_H37Rv S----------SFQYVTLVGGHVLAQFTLLVILAVFTRE--QVHEFGWRI
Mb3503c|M.bovis_AF2122/97 S----------SFQYVTLVGGHVLAQFTLLVILAVFTRE--QVHEFGWRI
MAB_3808c|M.abscessus_ATCC_199 S----------SFQYVTLVGGHVLAQFTLLIILTVFDTA--QVHEFGWRI
MSMEG_0985|M.smegmatis_MC2_155 S----------SFQYVTLVGGHVLAQFTLLIILTAFTTE--QVHDFGWRI
MMAR_4238|M.marinum_M G----------MFTVLGGGVALVVSGLTFLGVNYTIGESSSAFLQWGWRV
MUL_0949|M.ulcerans_Agy99 G----------MFTVLGGGVALVVSGLTFLGVNYTIGESSSALLQWGWRV
MAV_1344|M.avium_104 G----------MFTLLGGGTAGILASLTFLAVNLTMGEHSPAFMHWGWRV
Mflv_2381|M.gilvum_PYR-GCK V----------SFLVWSVVVGFLAGAITVTALETWLSED--AMNSYGWRI
Mvan_4266|M.vanbaalenii_PYR-1 V----------SFLVWSVVVGFLLGSVTVTALEATLSES--AMNSFGWRI
TH_3400|M.thermoresistible__bu G----------SFLEFGTLAGFSLGALMMLGVSLMLSDD--QMNTWGWRV
MLBr_01562|M.leprae_Br4923 VGGALTEVSWRLAFLVNVPIGLVMIYLARTALHETHKERMKLDAAGAILA
. . : .
Rv3476c|M.tuberculosis_H37Rv GFAVGGGAAIVVF-----WLRRTMDESLSQERLTAIKAG---RDHDSGSL
Mb3503c|M.bovis_AF2122/97 GFAVGGGAAIVVF-----WLRRTMDESLSQERLTAIKAG---RDHDSGSL
MAB_3808c|M.abscessus_ATCC_199 AFATGGVAAIVVY-----WLRRTMDESLSEEQLAAIKAG---ADTSSGSM
MSMEG_0985|M.smegmatis_MC2_155 GFAIGGVAAIVVF-----WLRRTMDESLSEEQLEAIKTG---KDTSSGSL
MMAR_4238|M.marinum_M PFLLSALLIAFAL-----YVRLEINETPVFARAMAQADDQESTAAARAPL
MUL_0949|M.ulcerans_Agy99 PFLLSALLIAFAL-----YVRLEINETPVFARAMAQADDQESTAAARAPL
MAV_1344|M.avium_104 PFLISSGLIGIAL-----YVRLNIDETPIFVEEKARH------LVPKAPL
Mflv_2381|M.gilvum_PYR-GCK PFMIAGLLGAVGL-----YIRLKLGDTPEFRSLRESG---EVSQ---SPL
Mvan_4266|M.vanbaalenii_PYR-1 PFLIAGALGAVGL-----YIRLRLGDTPEFEALRESG---EVST---TPL
TH_3400|M.thermoresistible__bu PFLIAAPLGLIGL-----YLRSRLDDTPIFRELQESD---REEKSIWTEF
MLBr_01562|M.leprae_Br4923 TLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAENPVV
: . . :: : .
Rv3476c|M.tuberculosis_H37Rv RELATHYWKPLLLCFLVTLGGTVAFYTYSVNAPAIVKSVYGSQAMTATWI
Mb3503c|M.bovis_AF2122/97 RELATHYWKPLLLCFLVTLGGTVAFYTYSVNAPAIVKSVYGSQAMTATWI
MAB_3808c|M.abscessus_ATCC_199 RELLTRYRKPLLLCFLITMGGTLAFYTYSVNAPAIVKAAYKDQAMTATWI
MSMEG_0985|M.smegmatis_MC2_155 GELLTRYPKALLMCFLITLGGTIAFYTYSVNAPAIVKTTYKDQAMTATWI
MMAR_4238|M.marinum_M AAVLCRQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLI
MUL_0949|M.ulcerans_Agy99 AAVLCRQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLI
MAV_1344|M.avium_104 TELLRLQRREIILVAGSFVGGMGFIYLGNTFLVMYAHNHLGYSRSFIWGI
Mflv_2381|M.gilvum_PYR-GCK REAVTTSWRPILQIIGLVVIHNVGFYIVYTFLPSYFTETLDFSKVDAFWS
Mvan_4266|M.vanbaalenii_PYR-1 KEAVTTSWRPILQIIGLVVVHNVGFYVVYTFLPSYFTRTLEFSKIDAFYS
TH_3400|M.thermoresistible__bu KDLLVEYRGQILRLGGLVVALNVVNYTLLSYMPTYLQTSIGLSTDTALVV
MLBr_01562|M.leprae_Br4923 PFDLFRDRNRLVTFIAIFLAG-GVIFSLTVSIGLYIQDILGYSALRAGIG
:: : : .
Rv3476c|M.tuberculosis_H37Rv NLVGLILLMMLQPIGGMISDKIGRKPLLLWF---------------GVGG
Mb3503c|M.bovis_AF2122/97 NLVGLILLMMLQPIGGMISDKIGRKPLLLWF---------------GVGG
MAB_3808c|M.abscessus_ATCC_199 NLAGLIFLMLLQPVGGIISDKVGRKPLLLFF---------------GFGG
MSMEG_0985|M.smegmatis_MC2_155 NLFGLIFLMLLQPVGGLISDKVGRKPLLLWF---------------GFGG
MMAR_4238|M.marinum_M GVLGGLVCIGFVALSAILCDRFGRRRVMLVG---------------WAVG
MUL_0949|M.ulcerans_Agy99 GVLGGLVCIGFVALSAILCDRFGRRRVMLVG---------------WAVG
MAV_1344|M.avium_104 GALGGLTSMACVACSAWISDRVGRRRVMLWG---------------LVAC
Mflv_2381|M.gilvum_PYR-GCK ITVASLVGIVLIPPLGALSDRIGRKSLLIAG---------------AVAF
Mvan_4266|M.vanbaalenii_PYR-1 ITVASLVGIVLIPPLGALSDRVGRKPLLLAG---------------SIGF
TH_3400|M.thermoresistible__bu PIIGMLSMMVFLPFAGLLSDRFGRKPMWWIS---------------LGGL
MLBr_01562|M.leprae_Br4923 FIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMNRGV
: . : .:.. : :
Rv3476c|M.tuberculosis_H37Rv LIYTYVLVTYLPETRSPTMSFLLVAVGYVILTGYCSINALVKSELFPAHV
Mb3503c|M.bovis_AF2122/97 LIYTYVLVTYLPETRSPTMSFLLVAVGYVILTGYCSINALVKSELFPAHV
MAB_3808c|M.abscessus_ATCC_199 VVYTYVLITYLPQVHAPIVSFLLVAVSYVLLTGYTSINALVKSELFPSHV
MSMEG_0985|M.smegmatis_MC2_155 VIYTYILITYLPQTHSAIMSFLLVAGGYIILTGYTSINAVVKAELFPSHI
MMAR_4238|M.marinum_M LFWSPLVMPLIGSGDTVGFTVALLGLYAVAASGFGPAASLIP-ELFATRY
MUL_0949|M.ulcerans_Agy99 LFWSPLVMPLIGSGDTVGFTVALLGLYAVAASGFGPAASLIP-ELFATRY
MAV_1344|M.avium_104 LPWAFVVIPLIDTGRPVCYVVAVLGMFGTAAVANGPTAAFVP-ELFATRY
Mflv_2381|M.gilvum_PYR-GCK AVFAYPLFLLLNTGSLAAAIAAHAALAAIESVFVSASLAAGA-ELFATKV
Mvan_4266|M.vanbaalenii_PYR-1 AVFAYPLFLLLNTGSLAAAIAAHAALAAIEAVFVCASLAAGA-ELFATRV
TH_3400|M.thermoresistible__bu LALVVPMFMLMST-SVPGAIIGFAVLGLLYVPQLATISATFP-AMFPTHV
MLBr_01562|M.leprae_Br4923 PYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIALMLQSL
: : . ::
Rv3476c|M.tuberculosis_H37Rv R---ALGVGVGYALANSVFGGTAPLIYQALKERDQVPMFIAYVTACIAVS
Mb3503c|M.bovis_AF2122/97 R---ALGVGVGYALANSVFGGTAPLIYQALKERDQVPMFIAYVTACIAVS
MAB_3808c|M.abscessus_ATCC_199 R---ALGVGVGYALANSVFGGTAPLIYQALKEHDQVPLFIGYVTVCIAIS
MSMEG_0985|M.smegmatis_MC2_155 R---ALGVGVGYALANSLVGGTAPVIYQALKERDQVPLFIGYVTVCIAIS
MMAR_4238|M.marinum_M R---YTGTALALNVAGIVGGAAPPLIAGTLLASYG-SSAVGLMMTALVAA
MUL_0949|M.ulcerans_Agy99 R---YTGTALALNVAGIVGGAAPPLIAGTLLASYG-SSAVGLMMTALVAA
MAV_1344|M.avium_104 R---YSGAAVAMNLAGIVGAAVPPLLAGTLLATYG-SWAIGLMMASLVLA
Mflv_2381|M.gilvum_PYR-GCK R---SSGYSIGYNVSVAVFGGTAPYVATWLVSRTGNDIAPAYYLIAAAIV
Mvan_4266|M.vanbaalenii_PYR-1 R---SSGYSIGYNVSVAIFGGTAPYVATWLVSRTGNDIAPAFYVIAAAVV
TH_3400|M.thermoresistible__bu R---YAGFAIAYNVSTSLFGGTAPAANDWLIQQTGNNLMPAFYMMAACLV
MLBr_01562|M.leprae_Br4923 GGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRALDHGYTYGLLWVA
. . : : . .. : .
Rv3476c|M.tuberculosis_H37Rv LIVYVFFIKNKADTYLDREQGFAFYGHA-----------
Mb3503c|M.bovis_AF2122/97 LIVYVFFIKNKADTYLDREQGFAFYGHA-----------
MAB_3808c|M.abscessus_ATCC_199 LVVYLFFLKNKSQTYLDREQGSAFNR-------------
MSMEG_0985|M.smegmatis_MC2_155 LVVYLFYLKNKSETYLDREKGAAFVR-------------
MMAR_4238|M.marinum_M SLLCTYLLPETAGAPLTAA--------------------
MUL_0949|M.ulcerans_Agy99 SLLCTYLLPETAGAPLTAA--------------------
MAV_1344|M.avium_104 SFVSVYLLPETRGAALDAAAAAEKVAAR-----------
Mflv_2381|M.gilvum_PYR-GCK SLLTVLTMRETARKPLR----------------KLVHP-
Mvan_4266|M.vanbaalenii_PYR-1 TFLTVLTMRETARQPLR----------------KLVSD-
TH_3400|M.thermoresistible__bu GMLALVKLPETARCPIGGTEIPGTEDAPPPVEYDLVGNR
MLBr_01562|M.leprae_Br4923 GAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL----
: :