For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSRVALACVVGSTVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATAFLSRPLGAAVFGHFGD RLGRKKTLVATLLIMALSTVTVGVVPSTATIGVSAPLILVALRLLQGFAVGGEWAGSALLSIESAPAHKR GYYGMFTVLGGGVALVVSGLTFLGVNYTIGESSSALLQWGWRVPFLLSALLIAFALYVRLEINETPVFAR AMAQADDQESTAAARAPLAAVLCRQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLIGV LGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGSGDTVGFTVALLGLYAVAASGFGPA ASLIPELFATRYRYTGTALALNVAGIVGGAAPPLIAGTLLASYGSSAVGLMMTALVAASLLCTYLLPETA GAPLTAA
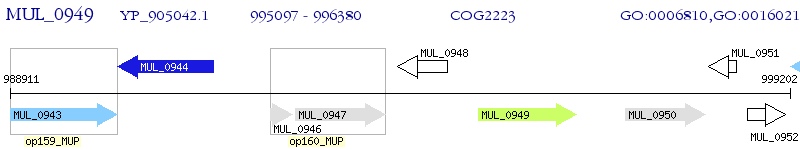
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0949 | - | - | 100% (427) | integral membrane transport protein |
| M. ulcerans Agy99 | MUL_3187 | - | 9e-17 | 27.02% (396) | sugar transporter |
| M. ulcerans Agy99 | MUL_2955 | - | 5e-14 | 23.92% (301) | fusion protein of transposase for IS2606 and sialic |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1232 | - | 1e-167 | 68.54% (426) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_2383 | - | 8e-79 | 40.58% (414) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv1200 | - | 1e-167 | 68.54% (426) | integral membrane transport protein |
| M. leprae Br4923 | MLBr_01562 | - | 4e-09 | 24.51% (355) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_3416 | - | 1e-106 | 48.44% (417) | membrane transporter |
| M. marinum M | MMAR_4238 | - | 0.0 | 99.77% (427) | integral membrane transport protein |
| M. avium 104 | MAV_1344 | - | 1e-148 | 62.53% (427) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_4058 | - | 1e-81 | 40.84% (431) | major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_3714 | - | 7e-78 | 40.43% (423) | PUTATIVE major facilitator superfamily MFS_1 |
| M. vanbaalenii PYR-1 | Mvan_2899 | - | 2e-76 | 38.14% (430) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0949|M.ulcerans_Agy99 ----------------------------------------------MSRV
MMAR_4238|M.marinum_M ----------------------------------------------MSRV
Mb1232|M.bovis_AF2122/97 ----------------------------------------------MKRV
Rv1200|M.tuberculosis_H37Rv ----------------------------------------------MKRV
MAV_1344|M.avium_104 --------------------------------MGEPSARATTPGTPMRRI
MAB_3416|M.abscessus_ATCC_1997 -----------------------------MTSSPAEPRQMADGQVSPRRV
Mflv_2383|M.gilvum_PYR-GCK ---------------------------------MGPGGRT---RVSPRRV
TH_3714|M.thermoresistible__bu ---------------------------------MSAEAATPTSRVTPRRV
Mvan_2899|M.vanbaalenii_PYR-1 ---------------------------------MNEASVAD--SALLRRV
MSMEG_4058|M.smegmatis_MC2_155 ---------------------------------MNRMQSS-------RKA
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
:
MUL_0949|M.ulcerans_Agy99 ALACVVGSTVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATA
MMAR_4238|M.marinum_M ALACVVGSTVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATA
Mb1232|M.bovis_AF2122/97 ALACLVGSAIEFYDFLIYGTAAALVFPTVFFPHLDPTVAAVASMGTFAVA
Rv1200|M.tuberculosis_H37Rv ALACLVGSAIEFYDFLIYGTAAALVFPTVFFPHLDPTVAAVASMGTFAVA
MAV_1344|M.avium_104 AAACLVGSAIEFYDFLIYGTAAALVFPAVFFPRLGPTVATIASMATFATA
MAB_3416|M.abscessus_ATCC_1997 FMASAVGSAIEFYDFYIYGTAAALVFPAVFFPNLSHSLALFASIATFSTA
Mflv_2383|M.gilvum_PYR-GCK AIASFVGTTVEFYDFLIYGTAAALVFPKLFFPNASPAAGILLSFATFGVG
TH_3714|M.thermoresistible__bu AVASFIGTTVEFYDFLIYGTAAALVFPKLFFPTASPASGVLLSFATFGVG
Mvan_2899|M.vanbaalenii_PYR-1 ALSSLLGTAIEYYDFLVYGTMSALVFGLVFFPDSDPAVATIAAFGTLAAG
MSMEG_4058|M.smegmatis_MC2_155 GLASLVGTTIEWYDFQIYGLSAAVVFSTVFFPEVSTLAGTLASFAVFALG
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVA-IVALPKIQNELSLSDAGRSWGIT
. :.:.: .: : . : * : * . : .
MUL_0949|M.ulcerans_Agy99 FLSRPLGAAVFG--HFGDRLGRKKTLVATLLIMALSTVTVGVVPSTATIG
MMAR_4238|M.marinum_M FLSRPLGAAVFG--HFGDRLGRKKTLVATLLIMALSTVTVGVVPSTATIG
Mb1232|M.bovis_AF2122/97 FLSRPFGAAVFG--YFGDRLGRKKTLVATLLIMGLATVTVGLVPTTVAIG
Rv1200|M.tuberculosis_H37Rv FLSRPFGAAVFG--YFGDRLGRKKTLVATLLIMGLATVTVGLVPTTVAIG
MAV_1344|M.avium_104 FLSRPLGAAVFG--YFGDRLGRKKTLVATLLIMGASTVSVGLVPSTASIG
MAB_3416|M.abscessus_ATCC_1997 FLARPIGAALFG--HYGDRFSRKGTLVVTLITMGLATMAVGLVPGAATIG
Mflv_2383|M.gilvum_PYR-GCK FIARPLGGIVFG--HFGDRVGRRRMLVYSLVGMGVSTVLIGVLPTYAQIG
TH_3714|M.thermoresistible__bu FFARPLGGIVFG--HFGDRLGRKRMLVYSLVGMGVATVLIGLLPTYAQIG
Mvan_2899|M.vanbaalenii_PYR-1 YVARPVGGIVFG--HFGDRIGRKSMLVLTMTLMGTASFLIGLLPSFAAVG
MSMEG_4058|M.smegmatis_MC2_155 FLARPLGGIIFG--HFGDRAGRKQILLITLLLMGISTFAVGVIPSYETIG
MLBr_01562|M.leprae_Br4923 AYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLV
..*. :: ** .*: :: : : ::. .: :
MUL_0949|M.ulcerans_Agy99 VS-------------APLILVALRLLQG-------FAVGGEWAGSALLSI
MMAR_4238|M.marinum_M VS-------------APLILVALRLLQG-------FAVGGEWAGSALLSI
Mb1232|M.bovis_AF2122/97 AA-------------APLILTTMRLLQG-------FAVGGEWAGSALLSA
Rv1200|M.tuberculosis_H37Rv AA-------------APLILTTMRLLQG-------FAVGGEWAGSALLSA
MAV_1344|M.avium_104 IA-------------APLLLTVLRLLQG-------FAVGAEWAGSVLLSA
MAB_3416|M.abscessus_ATCC_1997 AA-------------APTAIVILRLVQG-------LAVGGEWSGAALLSA
Mflv_2383|M.gilvum_PYR-GCK LA-------------APILLTVLRLAQG-------FAVGGEWGGATLMAV
TH_3714|M.thermoresistible__bu MA-------------APALLTLLRLAQG-------FAMGGEWGGATLMAV
Mvan_2899|M.vanbaalenii_PYR-1 VA-------------APILLVALRVIQG-------VAIGGEWGGATLMVT
MSMEG_4058|M.smegmatis_MC2_155 AW-------------APILLVAMRLLGG-------IALGGEWGGAVLLSV
MLBr_01562|M.leprae_Br4923 IARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGL
: :. . * .... . . ::
MUL_0949|M.ulcerans_Agy99 ESAPAHKRGYYGMFTVLGGGVALVVSGLTFLGVNYTIGESSSALLQWGWR
MMAR_4238|M.marinum_M ESAPAHKRGYYGMFTVLGGGVALVVSGLTFLGVNYTIGESSSAFLQWGWR
Mb1232|M.bovis_AF2122/97 EYAPASKRGWYGMFTVVGGGIALVLTSLTFLGVNYTIGESSPTFMQWGWR
Rv1200|M.tuberculosis_H37Rv EYAPASKRGWYGMFTVVGGGIALVLTSLTFLGVNYTIGESSPTFMQWGWR
MAV_1344|M.avium_104 EYAPTDRRGWYGMFTLLGGGTAGILASLTFLAVNLTMGEHSPAFMHWGWR
MAB_3416|M.abscessus_ATCC_1997 EYSPSQNRGRAATAVPIGTSTGLLLSSLVFLIVSLTIGEDSPAFLSWGWR
Mflv_2383|M.gilvum_PYR-GCK EHATADRRGFYGAFPQMGAPAQTAAATLAFYLAARLPDD---QFLAWGWR
TH_3714|M.thermoresistible__bu EHASADRKGFYGAFPQMGAPAGTATATLAFFAVSRLPDG---QFLSWGWR
Mvan_2899|M.vanbaalenii_PYR-1 EHAEAHRRGFWNGVMQMGSPIGSLLSVAVVTVITLLPDE---SFLSWGWR
MSMEG_4058|M.smegmatis_MC2_155 EHAPPGKRGFYGALPQVGSPAGFLLATTVFAIVTAATGD---AFETWGWR
MLBr_01562|M.leprae_Br4923 VVGGALTEVSWRLAFLVNVPIGLVMIYLARTALHETHKERMKLDAAGAIL
. . . :. . .
MUL_0949|M.ulcerans_Agy99 VPFLLSALLIAFAL-----YVRLEINETPVFARAMAQADDQESTAAARAP
MMAR_4238|M.marinum_M VPFLLSALLIAFAL-----YVRLEINETPVFARAMAQADDQESTAAARAP
Mb1232|M.bovis_AF2122/97 IPFLVSAALIAVAL-----YVRFNIDETPVFAR--ERADEKTRLGPAETP
Rv1200|M.tuberculosis_H37Rv IPFLVSAALIAVAL-----YVRFNIDETPVFAR--ERADEKTRLGPAETP
MAV_1344|M.avium_104 VPFLISSGLIGIAL-----YVRLNIDETPIFVE------EKARHLVPKAP
MAB_3416|M.abscessus_ATCC_1997 VPFIASGLLVAVGL-----YIRLQIAETPQFVE------AKAHHQLVKSP
Mflv_2383|M.gilvum_PYR-GCK IPFLVSAILIVIGL-----VVRLTVTESPHFAE----LQRRSE--VTRTP
TH_3714|M.thermoresistible__bu LPFLLSAVLIGIGL-----AVRLTLTESPDFAA----VQARSA--VHRMP
Mvan_2899|M.vanbaalenii_PYR-1 IPFLVSLLLLAVGI-----YVRLSVTESPVFEA----ARKEAAGNAPRVP
MSMEG_4058|M.smegmatis_MC2_155 IPFLLSGVLVAVGL-----FIRLAVPETPVFAE----IREKVQH-TASAP
MLBr_01562|M.leprae_Br4923 ATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAENPV
.: : :. ..: : : : .
MUL_0949|M.ulcerans_Agy99 LAAVLCRQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLL
MMAR_4238|M.marinum_M LAAVLCRQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLL
Mb1232|M.bovis_AF2122/97 IAQVLRRQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLGYSRGSILF
Rv1200|M.tuberculosis_H37Rv IAQVLRRQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLGYSRGSILF
MAV_1344|M.avium_104 LTELLRLQRREIILVAGSFVGGMGFIYLGNTFLVMYAHNHLGYSRSFIWG
MAB_3416|M.abscessus_ATCC_1997 LRHVITESPRTLVLTSGTMLIIFTLSFMTNTYFPGFARSELHFSRAAILA
Mflv_2383|M.gilvum_PYR-GCK IVAAFRRHWRQILLVAGAYLSQGIFAYICVAYLVSYATTVAEIPRDWALL
TH_3714|M.thermoresistible__bu IAEAFRRHRRQILLVAGAYLSQGVFAYICVAYLVSYGTSVAGIDRTATLF
Mvan_2899|M.vanbaalenii_PYR-1 FVEILRRP-RTLVLACAVGIGPFALTALISTYMISYATAIG-YHRTDVMT
MSMEG_4058|M.smegmatis_MC2_155 LIEVLRNHPKTVMLGIGAFAVSSGGYYVYATYMVAYGKNDLGISATTMLI
MLBr_01562|M.leprae_Br4923 VPFDLFRDRNRLVTFIAIFLAG-GVIFSLTVSIGLYIQDILGYSALRAGI
. : . :: . . : :
MUL_0949|M.ulcerans_Agy99 IGVLGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGSG
MMAR_4238|M.marinum_M IGVLGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGSG
Mb1232|M.bovis_AF2122/97 DSVLGGLLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLLVMPLIDSG
Rv1200|M.tuberculosis_H37Rv DSVLGGLLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLLVMPLIDSG
MAV_1344|M.avium_104 IGALGGLTSMACVACSAWISDRVGRRRVMLWGLVACLPWAFVVIPLIDTG
MAB_3416|M.abscessus_ATCC_1997 AGALGAVVLMVSATAGGILSDRLGRRTVILSSMTLTLPWTLLVTPLLTSG
Mflv_2383|M.gilvum_PYR-GCK GVFLGAVVAVATYPLFGALSDRVGRKPLYLAGVIAMALAVVPTFALINTG
TH_3714|M.thermoresistible__bu GVSVAAVLAVVMYPLFGSLSDTFGRKPLFLAGVIAMGVSIGPAFALIDTG
Mvan_2899|M.vanbaalenii_PYR-1 ALIFTSLTGLIAIPFFSALSDHVGRRLVMVAGAIGIIGYAWPFYALVDM-
MSMEG_4058|M.smegmatis_MC2_155 GGIVFACCAIVGNLTSASVSDRIGRRPVFLTMALFTILYAFPLFWLVETE
MLBr_01562|M.leprae_Br4923 GFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMNRG
. : . .. : : : ::
MUL_0949|M.ulcerans_Agy99 DTVGF-TVALLGLYAVA-ASGFGPAASLIPELFATRYRYTGTALALNVAG
MMAR_4238|M.marinum_M DTVGF-TVALLGLYAVA-ASGFGPAASLIPELFATRYRYTGTALALNVAG
Mb1232|M.bovis_AF2122/97 SPSLF-AVAVVGMYAIG-GFGFGPTASFIPELFATSYRYTGSALAANLAG
Rv1200|M.tuberculosis_H37Rv SPSLF-AVAVVGMYAIG-GFGFGPTASFIPELFATSYRYTGSALAANLAG
MAV_1344|M.avium_104 RPVCY-VVAVLGMFGTA-AVANGPTAAFVPELFATRYRYSGAAVAMNLAG
MAB_3416|M.abscessus_ATCC_1997 PFWAY-LAAMALTYVLF-GLAFGPMASFLPESYPVGTRYSGTGVAFNVAG
Mflv_2383|M.gilvum_PYR-GCK RPVMF-VLALVLIFGLAMAPAAGVTGALFSMAFDADVRYSGVSVGYTLSQ
TH_3714|M.thermoresistible__bu RPALF-LVALVLVFGLAMAPAAGVTGALFAMTFDADVRYSGVSIGYTLSQ
Mvan_2899|M.vanbaalenii_PYR-1 RSQAY-LTAAMVIAQLIQSMMYAPLGALFSEMFGTTVRYTGASMGYQLAA
MSMEG_4058|M.smegmatis_MC2_155 VAVLI-WIAQG-IGGFANGSLYGLMGSLSAEMFEPHIRYTGASLGQQVSA
MLBr_01562|M.leprae_Br4923 VPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIALMLQS
* . . .:. :
MUL_0949|M.ulcerans_Agy99 IVGGAAPPLIAGTLLASYG-SSAVG--------------------LMMTA
MMAR_4238|M.marinum_M IVGGAAPPLIAGTLLASYG-SSAVG--------------------LMMTA
Mb1232|M.bovis_AF2122/97 VAGGALPPVIAGALVATYG-SWAIG--------------------VMLAI
Rv1200|M.tuberculosis_H37Rv VAGGALPPVIAGALVATYG-SWAIG--------------------VMLAI
MAV_1344|M.avium_104 IVGAAVPPLLAGTLLATYG-SWAIG--------------------LMMAS
MAB_3416|M.abscessus_ATCC_1997 VFGGAIPPLIAGPLQNTLG-SWAVG--------------------LMLLV
Mflv_2383|M.gilvum_PYR-GCK VLGSAFAPTIATALYAATGSSNSIA--------------------AYLIA
TH_3714|M.thermoresistible__bu VLGSAFAPTIAAALYAATESSGSIV--------------------AYLIG
Mvan_2899|M.vanbaalenii_PYR-1 LLGAGFTPLIASALHAQEVSRTPLV--------------------ALAAG
MSMEG_4058|M.smegmatis_MC2_155 GLVGGTTPMIVTAMVASAGHSWPAP--------------------AWLAL
MLBr_01562|M.leprae_Br4923 LGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRALDHGYTYGLLWV
. : :
MUL_0949|M.ulcerans_Agy99 LVAASLLCTYLLP-----------ETAGAPLTAA---------
MMAR_4238|M.marinum_M LVAASLLCTYLLP-----------ETAGAPLTAA---------
Mb1232|M.bovis_AF2122/97 LALISLVCTYRLP-----------ETAGSALVSR---------
Rv1200|M.tuberculosis_H37Rv LALISLVCTYRLP-----------ETAGSALVSR---------
MAV_1344|M.avium_104 LVLASFVSVYLLP-----------ETRGAALDAAAAAEKVAAR
MAB_3416|M.abscessus_ATCC_1997 ICAISVASISGMR-----------ETWAGSAR-----------
Mflv_2383|M.gilvum_PYR-GCK ASAVSAVAVCFMPGGWR---AAPAEARPEHTPTAPTG------
TH_3714|M.thermoresistible__bu VSVISAVSVCLLPGGWGRTGDQPATEVPASVRPMVSGPD----
Mvan_2899|M.vanbaalenii_PYR-1 CGLVTVVAVWRIS-----------ETRGTDLTTV---------
MSMEG_4058|M.smegmatis_MC2_155 LAAVSLLCVLVLPETYNKPIVAVEKGDATSSEAART-------
MLBr_01562|M.leprae_Br4923 AGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL-------
: