For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRVALACVVGSTVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATAFLSRPLGAAVFGHFGDR LGRKKTLVATLLIMALSTVTVGVVPSTATIGVSAPLILVALRLLQGFAVGGEWAGSALLSIESAPAHKRG YYGMFTVLGGGVALVVSGLTFLGVNYTIGESSSAFLQWGWRVPFLLSALLIAFALYVRLEINETPVFARA MAQADDQESTAAARAPLAAVLCRQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLIGVL GGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGSGDTVGFTVALLGLYAVAASGFGPAA SLIPELFATRYRYTGTALALNVAGIVGGAAPPLIAGTLLASYGSSAVGLMMTALVAASLLCTYLLPETAG APLTAA*
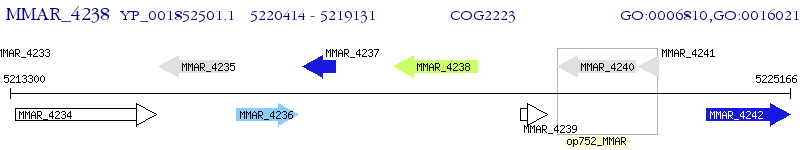
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4238 | - | - | 100% (427) | integral membrane transport protein |
| M. marinum M | MMAR_2574 | - | 2e-16 | 27.02% (396) | sugar transporter |
| M. marinum M | MMAR_2797 | nanT | 2e-14 | 23.60% (322) | sialic acid-transport integral membrane protein NanT |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1232 | - | 1e-168 | 68.78% (426) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_2383 | - | 2e-79 | 40.82% (414) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv1200 | - | 1e-168 | 68.78% (426) | integral membrane transport protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3416 | - | 1e-107 | 48.68% (417) | membrane transporter |
| M. avium 104 | MAV_1344 | - | 1e-149 | 62.76% (427) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_4058 | - | 2e-82 | 41.07% (431) | major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_3714 | - | 1e-78 | 40.66% (423) | PUTATIVE major facilitator superfamily MFS_1 |
| M. ulcerans Agy99 | MUL_0949 | - | 0.0 | 99.77% (427) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_2899 | - | 5e-77 | 38.37% (430) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2383|M.gilvum_PYR-GCK ----MGPGGRT---RVSPRRVAIASFVGTTVEFYDFLIYGTAAALVFPKL
TH_3714|M.thermoresistible__bu ----MSAEAATPTSRVTPRRVAVASFIGTTVEFYDFLIYGTAAALVFPKL
Mvan_2899|M.vanbaalenii_PYR-1 ----MNEASVAD--SALLRRVALSSLLGTAIEYYDFLVYGTMSALVFGLV
MSMEG_4058|M.smegmatis_MC2_155 ----MNRMQSS-------RKAGLASLVGTTIEWYDFQIYGLSAAVVFSTV
MMAR_4238|M.marinum_M -----------------MSRVALACVVGSTVEYYDFCIYGTAAALVFPAV
MUL_0949|M.ulcerans_Agy99 -----------------MSRVALACVVGSTVEYYDFCIYGTAAALVFPAV
Mb1232|M.bovis_AF2122/97 -----------------MKRVALACLVGSAIEFYDFLIYGTAAALVFPTV
Rv1200|M.tuberculosis_H37Rv -----------------MKRVALACLVGSAIEFYDFLIYGTAAALVFPTV
MAV_1344|M.avium_104 ---MGEPSARATTPGTPMRRIAAACLVGSAIEFYDFLIYGTAAALVFPAV
MAB_3416|M.abscessus_ATCC_1997 MTSSPAEPRQMADGQVSPRRVFMASAVGSAIEFYDFYIYGTAAALVFPAV
: :. :*:::*:*** :** :*:** :
Mflv_2383|M.gilvum_PYR-GCK FFPNASPAAGILLSFATFGVGFIARPLGGIVFGHFGDRVGRRRMLVYSLV
TH_3714|M.thermoresistible__bu FFPTASPASGVLLSFATFGVGFFARPLGGIVFGHFGDRLGRKRMLVYSLV
Mvan_2899|M.vanbaalenii_PYR-1 FFPDSDPAVATIAAFGTLAAGYVARPVGGIVFGHFGDRIGRKSMLVLTMT
MSMEG_4058|M.smegmatis_MC2_155 FFPEVSTLAGTLASFAVFALGFLARPLGGIIFGHFGDRAGRKQILLITLL
MMAR_4238|M.marinum_M FYPGLSPALATIASMGTFATAFLSRPLGAAVFGHFGDRLGRKKTLVATLL
MUL_0949|M.ulcerans_Agy99 FYPGLSPALATIASMGTFATAFLSRPLGAAVFGHFGDRLGRKKTLVATLL
Mb1232|M.bovis_AF2122/97 FFPHLDPTVAAVASMGTFAVAFLSRPFGAAVFGYFGDRLGRKKTLVATLL
Rv1200|M.tuberculosis_H37Rv FFPHLDPTVAAVASMGTFAVAFLSRPFGAAVFGYFGDRLGRKKTLVATLL
MAV_1344|M.avium_104 FFPRLGPTVATIASMATFATAFLSRPLGAAVFGYFGDRLGRKKTLVATLL
MAB_3416|M.abscessus_ATCC_1997 FFPNLSHSLALFASIATFSTAFLARPIGAALFGHYGDRFSRKGTLVVTLI
*:* . . . ::..:. .:.:**.*. :**::*** .*: *: ::
Mflv_2383|M.gilvum_PYR-GCK GMGVSTVLIGVLPTYAQIGLAAPILLTVLRLAQGFAVGGEWGGATLMAVE
TH_3714|M.thermoresistible__bu GMGVATVLIGLLPTYAQIGMAAPALLTLLRLAQGFAMGGEWGGATLMAVE
Mvan_2899|M.vanbaalenii_PYR-1 LMGTASFLIGLLPSFAAVGVAAPILLVALRVIQGVAIGGEWGGATLMVTE
MSMEG_4058|M.smegmatis_MC2_155 LMGISTFAVGVIPSYETIGAWAPILLVAMRLLGGIALGGEWGGAVLLSVE
MMAR_4238|M.marinum_M IMALSTVTVGVVPSTATIGVSAPLILVALRLLQGFAVGGEWAGSALLSIE
MUL_0949|M.ulcerans_Agy99 IMALSTVTVGVVPSTATIGVSAPLILVALRLLQGFAVGGEWAGSALLSIE
Mb1232|M.bovis_AF2122/97 IMGLATVTVGLVPTTVAIGAAAPLILTTMRLLQGFAVGGEWAGSALLSAE
Rv1200|M.tuberculosis_H37Rv IMGLATVTVGLVPTTVAIGAAAPLILTTMRLLQGFAVGGEWAGSALLSAE
MAV_1344|M.avium_104 IMGASTVSVGLVPSTASIGIAAPLLLTVLRLLQGFAVGAEWAGSVLLSAE
MAB_3416|M.abscessus_ATCC_1997 TMGLATMAVGLVPGAATIGAAAPTAIVILRLVQGLAVGGEWSGAALLSAE
*. ::. :*::* :* ** :. :*: *.*:*.**.*:.*: *
Mflv_2383|M.gilvum_PYR-GCK HATADRRGFYGAFPQMGAPAQTAAATLAFYLAARLPDD---QFLAWGWRI
TH_3714|M.thermoresistible__bu HASADRKGFYGAFPQMGAPAGTATATLAFFAVSRLPDG---QFLSWGWRL
Mvan_2899|M.vanbaalenii_PYR-1 HAEAHRRGFWNGVMQMGSPIGSLLSVAVVTVITLLPDE---SFLSWGWRI
MSMEG_4058|M.smegmatis_MC2_155 HAPPGKRGFYGALPQVGSPAGFLLATTVFAIVTAATGD---AFETWGWRI
MMAR_4238|M.marinum_M SAPAHKRGYYGMFTVLGGGVALVVSGLTFLGVNYTIGESSSAFLQWGWRV
MUL_0949|M.ulcerans_Agy99 SAPAHKRGYYGMFTVLGGGVALVVSGLTFLGVNYTIGESSSALLQWGWRV
Mb1232|M.bovis_AF2122/97 YAPASKRGWYGMFTVVGGGIALVLTSLTFLGVNYTIGESSPTFMQWGWRI
Rv1200|M.tuberculosis_H37Rv YAPASKRGWYGMFTVVGGGIALVLTSLTFLGVNYTIGESSPTFMQWGWRI
MAV_1344|M.avium_104 YAPTDRRGWYGMFTLLGGGTAGILASLTFLAVNLTMGEHSPAFMHWGWRV
MAB_3416|M.abscessus_ATCC_1997 YSPSQNRGRAATAVPIGTSTGLLLSSLVFLIVSLTIGEDSPAFLSWGWRV
: . .:* :* : .. . : ****:
Mflv_2383|M.gilvum_PYR-GCK PFLVSAILIVIGLVVRLTVTESPHFAE----LQRRSE--VTRTPIVAAFR
TH_3714|M.thermoresistible__bu PFLLSAVLIGIGLAVRLTLTESPDFAA----VQARSA--VHRMPIAEAFR
Mvan_2899|M.vanbaalenii_PYR-1 PFLVSLLLLAVGIYVRLSVTESPVFEA----ARKEAAGNAPRVPFVEILR
MSMEG_4058|M.smegmatis_MC2_155 PFLLSGVLVAVGLFIRLAVPETPVFAE----IREKVQH-TASAPLIEVLR
MMAR_4238|M.marinum_M PFLLSALLIAFALYVRLEINETPVFARAMAQADDQESTAAARAPLAAVLC
MUL_0949|M.ulcerans_Agy99 PFLLSALLIAFALYVRLEINETPVFARAMAQADDQESTAAARAPLAAVLC
Mb1232|M.bovis_AF2122/97 PFLVSAALIAVALYVRFNIDETPVFAR--ERADEKTRLGPAETPIAQVLR
Rv1200|M.tuberculosis_H37Rv PFLVSAALIAVALYVRFNIDETPVFAR--ERADEKTRLGPAETPIAQVLR
MAV_1344|M.avium_104 PFLISSGLIGIALYVRLNIDETPIFVE------EKARHLVPKAPLTELLR
MAB_3416|M.abscessus_ATCC_1997 PFIASGLLVAVGLYIRLQIAETPQFVE------AKAHHQLVKSPLRHVIT
**: * *: ..: :*: : *:* * . *: :
Mflv_2383|M.gilvum_PYR-GCK RHWRQILLVAGAYLSQGIFAYICVAYLVSYATTVAEIPRDWALLGVFLGA
TH_3714|M.thermoresistible__bu RHRRQILLVAGAYLSQGVFAYICVAYLVSYGTSVAGIDRTATLFGVSVAA
Mvan_2899|M.vanbaalenii_PYR-1 RP-RTLVLACAVGIGPFALTALISTYMISYATAIG-YHRTDVMTALIFTS
MSMEG_4058|M.smegmatis_MC2_155 NHPKTVMLGIGAFAVSSGGYYVYATYMVAYGKNDLGISATTMLIGGIVFA
MMAR_4238|M.marinum_M RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLIGVLGG
MUL_0949|M.ulcerans_Agy99 RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLIGVLGG
Mb1232|M.bovis_AF2122/97 RQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLGYSRGSILFDSVLGG
Rv1200|M.tuberculosis_H37Rv RQRREIVLAAGSAVCCFGFVYLASTYLASYAQTRLGYSRGSILFDSVLGG
MAV_1344|M.avium_104 LQRREIILVAGSFVGGMGFIYLGNTFLVMYAHNHLGYSRSFIWGIGALGG
MAB_3416|M.abscessus_ATCC_1997 ESPRTLVLTSGTMLIIFTLSFMTNTYFPGFARSELHFSRAAILAAGALGA
: ::* . : ::: :. . .
Mflv_2383|M.gilvum_PYR-GCK VVAVATYPLFGALSDRVGRKPLYLAGVIAMALAVVPTFALINTGRPVMFV
TH_3714|M.thermoresistible__bu VLAVVMYPLFGSLSDTFGRKPLFLAGVIAMGVSIGPAFALIDTGRPALFL
Mvan_2899|M.vanbaalenii_PYR-1 LTGLIAIPFFSALSDHVGRRLVMVAGAIGIIGYAWPFYALVDM-RSQAYL
MSMEG_4058|M.smegmatis_MC2_155 CCAIVGNLTSASVSDRIGRRPVFLTMALFTILYAFPLFWLVETEVAVLIW
MMAR_4238|M.marinum_M LVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGSGDTVGFT
MUL_0949|M.ulcerans_Agy99 LVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGSGDTVGFT
Mb1232|M.bovis_AF2122/97 LLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLLVMPLIDSGSPSLFA
Rv1200|M.tuberculosis_H37Rv LLCIVFTALSSALCDQLGRRRVLLAGWAVALPWSLLVMPLIDSGSPSLFA
MAV_1344|M.avium_104 LTSMACVACSAWISDRVGRRRVMLWGLVACLPWAFVVIPLIDTGRPVCYV
MAB_3416|M.abscessus_ATCC_1997 VVLMVSATAGGILSDRLGRRTVILSSMTLTLPWTLLVTPLLTSGPFWAYL
: . :.* .**: : : *:
Mflv_2383|M.gilvum_PYR-GCK LALVLIFGLAMAPAAGVTGALFSMAFDADVRYSGVSVGYTLSQVLGSAFA
TH_3714|M.thermoresistible__bu VALVLVFGLAMAPAAGVTGALFAMTFDADVRYSGVSIGYTLSQVLGSAFA
Mvan_2899|M.vanbaalenii_PYR-1 TAAMVIAQLIQSMMYAPLGALFSEMFGTTVRYTGASMGYQLAALLGAGFT
MSMEG_4058|M.smegmatis_MC2_155 IAQG-IGGFANGSLYGLMGSLSAEMFEPHIRYTGASLGQQVSAGLVGGTT
MMAR_4238|M.marinum_M VALLGLYAVA-ASGFGPAASLIPELFATRYRYTGTALALNVAGIVGGAAP
MUL_0949|M.ulcerans_Agy99 VALLGLYAVA-ASGFGPAASLIPELFATRYRYTGTALALNVAGIVGGAAP
Mb1232|M.bovis_AF2122/97 VAVVGMYAIG-GFGFGPTASFIPELFATSYRYTGSALAANLAGVAGGALP
Rv1200|M.tuberculosis_H37Rv VAVVGMYAIG-GFGFGPTASFIPELFATSYRYTGSALAANLAGVAGGALP
MAV_1344|M.avium_104 VAVLGMFGTA-AVANGPTAAFVPELFATRYRYSGAAVAMNLAGIVGAAVP
MAB_3416|M.abscessus_ATCC_1997 AAMALTYVLF-GLAFGPMASFLPESYPVGTRYSGTGVAFNVAGVFGGAIP
* . . .:: . : **:* .:. :: .. .
Mflv_2383|M.gilvum_PYR-GCK PTIATALYAATGSSNSIAAYLIAASAVSAVAVCFMPGGWR---AAPAEAR
TH_3714|M.thermoresistible__bu PTIAAALYAATESSGSIVAYLIGVSVISAVSVCLLPGGWGRTGDQPATEV
Mvan_2899|M.vanbaalenii_PYR-1 PLIASALHAQEVSRTPLVALAAGCGLVTVVAVWRIS-----------ETR
MSMEG_4058|M.smegmatis_MC2_155 PMIVTAMVASAGHSWPAPAWLALLAAVSLLCVLVLPETYNKPIVAVEKGD
MMAR_4238|M.marinum_M PLIAGTLLASYG-SSAVGLMMTALVAASLLCTYLLP-----------ETA
MUL_0949|M.ulcerans_Agy99 PLIAGTLLASYG-SSAVGLMMTALVAASLLCTYLLP-----------ETA
Mb1232|M.bovis_AF2122/97 PVIAGALVATYG-SWAIGVMLAILALISLVCTYRLP-----------ETA
Rv1200|M.tuberculosis_H37Rv PVIAGALVATYG-SWAIGVMLAILALISLVCTYRLP-----------ETA
MAV_1344|M.avium_104 PLLAGTLLATYG-SWAIGLMMASLVLASFVSVYLLP-----------ETR
MAB_3416|M.abscessus_ATCC_1997 PLIAGPLQNTLG-SWAVGLMLLVICAISVASISGMR-----------ETW
* :. .: . : . :
Mflv_2383|M.gilvum_PYR-GCK PEHTPTAPTG------
TH_3714|M.thermoresistible__bu PASVRPMVSGPD----
Mvan_2899|M.vanbaalenii_PYR-1 GTDLTTV---------
MSMEG_4058|M.smegmatis_MC2_155 ATSSEAART-------
MMAR_4238|M.marinum_M GAPLTAA---------
MUL_0949|M.ulcerans_Agy99 GAPLTAA---------
Mb1232|M.bovis_AF2122/97 GSALVSR---------
Rv1200|M.tuberculosis_H37Rv GSALVSR---------
MAV_1344|M.avium_104 GAALDAAAAAEKVAAR
MAB_3416|M.abscessus_ATCC_1997 AGSAR-----------