For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GKRQESRERIEAAIVEIGRQHLVTEGAAGLSMRAVARDLGMVSSAVYRYVASRDELLTLLVVDAYTELAD AVAAAVADVSAAGARWRSQIRAIAHATRNWALDQPARWALLYGSPVPGYHAPAERTVVPGTRVVVALFAA VARGIAEGDIPATEETVAQPLSGDFARLRAEFGLHVDDATMARCFALWAGLIGAVSLEVFGQYGADTLTD PRQLFDLQIDSLTGALTN*
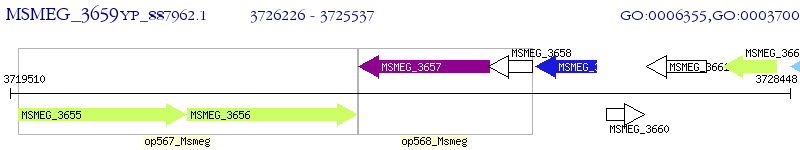
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3659 | - | - | 100% (229) | TetR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_2583 | - | 1e-07 | 29.17% (192) | putative transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6564 | - | 1e-05 | 34.69% (98) | transcriptional regulator, TetR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1846 | - | 4e-75 | 61.23% (227) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_3414 | - | 1e-82 | 67.84% (227) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1816 | - | 4e-75 | 61.23% (227) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4787c | - | 4e-22 | 32.75% (229) | TetR family transcriptional regulator |
| M. marinum M | MMAR_3679 | - | 8e-06 | 28.37% (208) | putative regulatory protein |
| M. avium 104 | MAV_2899 | - | 1e-80 | 65.35% (228) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2336 | - | 3e-08 | 30.54% (167) | transcriptional regulator, TetR family protein |
| M. ulcerans Agy99 | MUL_3622 | - | 1e-05 | 28.37% (208) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3167 | - | 2e-79 | 66.52% (221) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1846|M.bovis_AF2122/97 -------------MCQTCRVGKRRDAREQIEAKIVELGRRQLLDHGAAGL
Rv1816|M.tuberculosis_H37Rv -------------MCQTCRVGKRRDAREQIEAKIVELGRRQLLDHGAAGL
MAV_2899|M.avium_104 -------------------MGKRQQSREQIEARIIELGRRQLVDRGAAGL
MSMEG_3659|M.smegmatis_MC2_155 -------------------MGKRQESRERIEAAIVEIGRQHLVTEGAAGL
Mflv_3414|M.gilvum_PYR-GCK -------------------MGKRQDTRERIERDIVAIGRRHLVTEGAAGL
Mvan_3167|M.vanbaalenii_PYR-1 -------------MCDTGRVGKRQDTRDRIERQIVEIGRAHLVTHGAAAL
MAB_4787c|M.abscessus_ATCC_199 --------MPTRTNAADVPATRRARQRAQTTDEIKALARRQLADGGTGAL
MMAR_3679|M.marinum_M --------MFTLRTMYLSTVWEGDVRARFTVSDIASAALAIVDQDGLSGL
MUL_3622|M.ulcerans_Agy99 MISIRTSYVFTLRTMYLSTVWEGDVRARFTVSDIASAALAIVDQDGLSGL
TH_2336|M.thermoresistible__bu ----VSGVGKPISREAALLTSRASAGTGPTRQRLIEVAIELFKQHSVAGT
. : . . . ..
Mb1846|M.bovis_AF2122/97 SLRAIARNLGMVSSAVYRYVSSRDELLTLLLVDAYSDLADTVDRARDDTV
Rv1816|M.tuberculosis_H37Rv SLRAIARNLGMVSSAVYRYVSSRDELLTLLLVDAYSDLADTVDRARDDTV
MAV_2899|M.avium_104 SVRAIARDLGMVSSAVYRYVSSRDELLTLLLVDAYSDLADTVDRAR-ETA
MSMEG_3659|M.smegmatis_MC2_155 SMRAVARDLGMVSSAVYRYVASRDELLTLLVVDAYTELADAVAAAVADVS
Mflv_3414|M.gilvum_PYR-GCK SLRAIARDLGMVSSAVYRYVASRDELLTLLLVDAYTDLADTVETARAAST
Mvan_3167|M.vanbaalenii_PYR-1 SLRAIARELGMVSSAVYRYVASRDELLTLLLVDAYGELADSVERARDAAA
MAB_4787c|M.abscessus_ATCC_199 SLRGIAREMGMAPAALFRYFETQAALITALCVDANGALAQAMSDGQKAAP
MMAR_3679|M.marinum_M SMRSLAAALGTGPMTLYNYVRDRRELEALVAD-------AVVADVKVPAP
MUL_3622|M.ulcerans_Agy99 SMRSLAAALGTGPMTLYNYVRDRRELEALVAD-------AVVADVKVPAP
TH_2336|M.thermoresistible__bu SLQMISDALGLTKSAIYHHFRTRDELLTAVMEPLISEVAEIVSAAEARRG
*:: :: :* :::.:. : * : : :
Mb1846|M.bovis_AF2122/97 ADS--WSDDVIAIARAVRGWAVTNPARWALLYGSPVPGYHAPPDRTAGVA
Rv1816|M.tuberculosis_H37Rv ADS--WSDDVIAIARAVRGWAVTNPARWALLYGSPVPGYHAPPDRTAGVA
MAV_2899|M.avium_104 GEQ--WSDDVIAIARATRRWAVEHPACWALLYGSPVPGYHAPPERTVAAG
MSMEG_3659|M.smegmatis_MC2_155 AAGARWRSQIRAIAHATRNWALDQPARWALLYGSPVPGYHAPAERTVVPG
Mflv_3414|M.gilvum_PYR-GCK GD---WREQLRALGHAIRAWAVRQPASWALLYGSPVPGYHAPRDRTVTPG
Mvan_3167|M.vanbaalenii_PYR-1 GD---WQDRLRAVAHATRAWAVHHPARWALLYGSPVPGYHAPREQTVGPG
MAB_4787c|M.abscessus_ATCC_199 ESD--SAARLRAAFVAARRWALAHPGDFALLSGTPIPGYAAQPAETGPAA
MMAR_3679|M.marinum_M TDD--WRADVHAIATAMWETMRQHPNAIPLVLTR-----RTVSASSYMVA
MUL_3622|M.ulcerans_Agy99 IDD--WRADVHAIATAMWETMRQHPNAIPLVLTR-----RTVWASSYMVA
TH_2336|M.thermoresistible__bu ART-----RADHFLRGYAALAAAHREVISLLTGDP---------------
. : .*:
Mb1846|M.bovis_AF2122/97 TRVVGAFFDAIAAGIATGDIRLTDDVAPQPMSS--DFEKIRQEFGFPGDD
Rv1816|M.tuberculosis_H37Rv TRVVGAFFDAIAAGIATGDIRLTDDVAPQPMSS--DFEKIRQEFGFPGDD
MAV_2899|M.avium_104 TRVVAALFDAVAAGITTGDIRLTNDPAPQPMSS--DFERIRHEFGFPGDD
MSMEG_3659|M.smegmatis_MC2_155 TRVVVALFAAVARGIAEGDIPATEETVAQPLSG--DFARLRAEFGLHVDD
Mflv_3414|M.gilvum_PYR-GCK TRVVAALFDVVSAGIAAGDIASGEHPAPQPLSG--DFDRLREQFGFAGDD
Mvan_3167|M.vanbaalenii_PYR-1 TRVVGLLFSAIAEGVAAGAVRTSKVTVPQSLSS--DFDRLRAEFEFVADD
MAB_4787c|M.abscessus_ATCC_199 MRVIASFMDAYLRAVAAGEAVPRRTRFRPSAPGPLLVAMFGEDTAPPIDP
MMAR_3679|M.marinum_M DRLIAALG---RSGVGDADLLASFRGVLGLVMG-SAQAELAGPLAGTGRE
MUL_3622|M.ulcerans_Agy99 DRLIAALG---RSGVGDADLLASFRGVLGLVMG-SAQAELAGPLAGTGRE
TH_2336|M.thermoresistible__bu ---------------GVVALLRTRPDWAEVVQR--QMALLGDVQPGPGGR
:
Mb1846|M.bovis_AF2122/97 RVVTKCFLLWAGVVGAISLEVFGQYGADMLTDPGVVFDAQTRLLVAVLAE
Rv1816|M.tuberculosis_H37Rv RVVTKCFLLWAGVVGAISLEVFGQYGADMLTDPGVVFDAQTRLLVAVLAE
MAV_2899|M.avium_104 LVIAKCFLLWAGVLGAISLEVFGQYGADTLTDVEAVFDAQLRLLVDVLTR
MSMEG_3659|M.smegmatis_MC2_155 ATMARCFALWAGLIGAVSLEVFGQYGADTLTDPRQLFDLQIDSLTGALTN
Mflv_3414|M.gilvum_PYR-GCK VLLGKCLTLWAALIGAVGLEVFGQYGPDTFADPALAFDTQIAALTEMLCQ
Mvan_3167|M.vanbaalenii_PYR-1 DVVARCLVLWAALVGAISLEVFGQYGADTFADASVVFDAQMDLVIDLFCR
MAB_4787c|M.abscessus_ATCC_199 AIIGIGIFAWASITGYLAGEVFGSLG-DLVADSDALYHDHVDTVLCGMGF
MMAR_3679|M.marinum_M KEQVAVAARIGGLAGAEHPHIAALAQASQRSTPAADFERALDILLAGIES
MUL_3622|M.ulcerans_Agy99 KEQVAVAARIGGLAGAEHPHIAALAQASQRSTPAADFERALDILLAGIES
TH_2336|M.thermoresistible__bu ---VKAAVVMAGIAAAAGVDYGDLDEAALRDELLAAGRRTLGLRAPHARG
..: . .
Mb1846|M.bovis_AF2122/97 H-------
Rv1816|M.tuberculosis_H37Rv H-------
MAV_2899|M.avium_104 H-------
MSMEG_3659|M.smegmatis_MC2_155 --------
Mflv_3414|M.gilvum_PYR-GCK --------
Mvan_3167|M.vanbaalenii_PYR-1 QGAN----
MAB_4787c|M.abscessus_ATCC_199 AGV-----
MMAR_3679|M.marinum_M FERRRDRR
MUL_3622|M.ulcerans_Agy99 FERRRDRR
TH_2336|M.thermoresistible__bu --------