For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGLFDLTGRLAVVTGSSRGIGLAIAAGLAEAGARVVLNGVDGPRLEHTCDGLREKFGTDAVYAARFDVTD PDEVSSAIDDIEQRIGPIEVLVNNAGLQHREPLLEVSLANWQRVIDVDLTSAFLVGREVARHQLARGSGK IINICSVQTDLARPSIAAYTAAKGGLRNLTRAMTAEWAGGGLQINGLAPGYIHTEMTQALVDDTSFNNWI LGRTPAGRWGTVDDLVGPAIFLASPASAYVNGQVLFVDGGMTAVV*
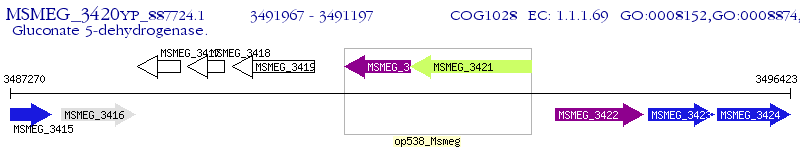
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3420 | - | - | 100% (256) | gluconate 5-dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3604 | - | 3e-40 | 35.94% (256) | sorbitol utilization protein SOU2 |
| M. smegmatis MC2 155 | MSMEG_4117 | - | 1e-39 | 35.48% (248) | 2-deoxy-D-gluconate 3-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1963c | - | 8e-33 | 32.81% (256) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0305 | - | 7e-42 | 40.24% (251) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1928c | - | 8e-33 | 32.81% (256) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 6e-35 | 35.39% (243) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_2723c | - | 1e-33 | 34.16% (243) | 3-oxoacyl- |
| M. marinum M | MMAR_2849 | - | 1e-35 | 33.59% (256) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_2775 | - | 2e-41 | 38.19% (254) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4266 | - | 3e-37 | 38.31% (248) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1491 | fabG1 | 2e-34 | 37.45% (243) | 3-oxoacyl- |
| M. vanbaalenii PYR-1 | Mvan_0894 | - | 8e-47 | 39.92% (253) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1963c|M.bovis_AF2122/97 -------------MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQ
Rv1928c|M.tuberculosis_H37Rv -------------MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQ
MMAR_2849|M.marinum_M -------------MSVLDLFDLSGKRALITGASTGIGKKVALAYAEAGAQ
MAV_2775|M.avium_104 ---------------MLDLFDLRGRRALVTGASTGIGKQVAQAYLQAGAR
Mflv_0305|M.gilvum_PYR-GCK -----------MTTTSANPFDLTGHVAVVTGGGSGIGLGMAGGLARAGAT
TH_4266|M.thermoresistible__bu ---------------VDDRFSMAGRVAVITGGGTGIGRASALVLAEYGAD
MSMEG_3420|M.smegmatis_MC2_155 ---------------MTGLFDLTGRLAVVTGSSRGIGLAIAAGLAEAGAR
MLBr_01807|M.leprae_Br4923 -MTDAAAKV--SAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHR
MAB_2723c|M.abscessus_ATCC_199 -MTNA---------------EFVPRSVLVTGGNRGIGLAVAQRLAADGHK
MUL_1491|M.ulcerans_Agy99 -MTDTATEQNTESAADGGKPAFVSRSVLVTGGNRGIGLAIAQRLATDGHR
Mvan_0894|M.vanbaalenii_PYR-1 MTVDIAREQPATAPRPDQMFDLTGRTALVTGASSGLGVQFALALRSAGAE
: : ::**.. *:* * *
Mb1963c|M.bovis_AF2122/97 VAIAARHLDALEKLADEI-GTSG-GKVVPVCCDVSQHQQVTSMLDQVTAE
Rv1928c|M.tuberculosis_H37Rv VAIAARHLDALEKLADEI-GTSG-GKVVPVCCDVSQHQQVTSMLDQVTAE
MMAR_2849|M.marinum_M VAVAARHSDALQVVADEI-AGVG-GKALPIRCDVTQPDQVRGMLDQMTGE
MAV_2775|M.avium_104 VAIAARDFEALQRTADEL-TAGGTGGVVAIRCDVTQPDEVSGMVDRMIAE
Mflv_0305|M.gilvum_PYR-GCK VAILGRNAQRLDDAAATL-RTHG-NPVLPLVCDVTDEEAVTETMARIRDE
TH_4266|M.thermoresistible__bu IVLAGRRPEPLKKTAAEI-EALG-RRALAVPTDVTAPDECRALVDATVGE
MSMEG_3420|M.smegmatis_MC2_155 VVLNGVDGPRLEHTCDGLREKFGTDAVYAARFDVTDPDEVSSAIDDIEQR
MLBr_01807|M.leprae_Br4923 VAVTHRASV-------VPDWFFG------VECDVTDNDAIGAAFKAVEEH
MAB_2723c|M.abscessus_ATCC_199 VAVTHRGSG-------APEGLLG------VQCDITDNDQIDAAFKEVEEK
MUL_1491|M.ulcerans_Agy99 VAVTHRGSG-------APEGLFG------VECDVTDNDAVDRAFKEVEEH
Mvan_0894|M.vanbaalenii_PYR-1 VVVAARRADRLAELCEAHEGLSA------VSCDVADPASIESAYQCVEER
:.: . *:: .
Mb1963c|M.bovis_AF2122/97 LGGIDIAVCNAGIIT-VTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAKAM
Rv1928c|M.tuberculosis_H37Rv LGGIDIAVCNAGIIT-VTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAKAM
MMAR_2849|M.marinum_M LGGIDIAVCNAGIVS-VQAMLDMPLEEFQRIQDTNVTGVFLTAQAAARAM
MAV_2775|M.avium_104 LGGIDIAVCNAGIIS-VSPMLEMPAAEFQRIQDTNVTGVFLTAQAAARAM
Mflv_0305|M.gilvum_PYR-GCK FGWLDSCFANAGVRGRFTPVLETSLAEFRSVTSVDLDGVFLTLREAARQM
TH_4266|M.thermoresistible__bu FGRLDVLVNNAGGGA-TKPLMKWTDDEWHEVLNLNLSSAWYLSRAAAKPM
MSMEG_3420|M.smegmatis_MC2_155 IGPIEVLVNNAGLQH-REPLLEVSLANWQRVIDVDLTSAFLVGREVARHQ
MLBr_01807|M.leprae_Br4923 QGPVEVLVANAGITS-DMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNM
MAB_2723c|M.abscessus_ATCC_199 QGPVEVLVANAGITA-DMLIMRMSEEQFLKVIDANLIGTFRLAKRASRNM
MUL_1491|M.ulcerans_Agy99 QGPVEVLVSNAGLSA-DAFLIRMTEERFEKVIDANLTGAFRVAQRASRSM
Mvan_0894|M.vanbaalenii_PYR-1 VGAVDILVNNAGTSI-PARAEDESLTDFQYVLDVNLVGAFRLTQLVGRKM
* :: . *** . : : . :: ..: : ..:
Mb1963c|M.bovis_AF2122/97 VKQGQGGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPH
Rv1928c|M.tuberculosis_H37Rv VKQGQGGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPH
MMAR_2849|M.marinum_M VDQGLGGTIITTASMSGHIINIPQQVSHYCTSKAAVVHLTKAMAVELAPH
MAV_2775|M.avium_104 VRQGRGGAIITTASMSGHIINVPQQVGHYCASKAAVIQLTKAMAVEFAPH
Mflv_0305|M.gilvum_PYR-GCK VAAGRGGSLVGVSSLG-AFQGMPRQP-AYAASKAGVTSMMDSFAVELARH
TH_4266|M.thermoresistible__bu IDQGRG--AIVNISSGAGLLAMPQAP-VYAAAKAGLNNLTGSMAAAWTRK
MSMEG_3420|M.smegmatis_MC2_155 LARGSG-KIINICSVQTDLARP--SIAAYTAAKGGLRNLTRAMTAEWAGG
MLBr_01807|M.leprae_Br4923 IKQRFG-RYIFMGSVVGLSGVP--GQANYAASKSGLIGMARSLARELGSR
MAB_2723c|M.abscessus_ATCC_199 IKKRFG-RLIFMGSVAGQIGLP--GQANYSASKAGLVGMARALTRELGSR
MUL_1491|M.ulcerans_Agy99 QRKKFG-RLIFIGSVSGSWGIG--NQANYAASKAGVIGMARSIARELSKV
Mvan_0894|M.vanbaalenii_PYR-1 IERGEG-SIVNVASILGMVAAAPNRQAAYCASKGGLINLTRELAVQWARY
* : * * ::*..: : ::
Mb1963c|M.bovis_AF2122/97 KIRVNSVSPGYILTELVEP---YTEYQPLWEPKIP-LGRLGRPEELA--G
Rv1928c|M.tuberculosis_H37Rv KIRVNSVSPGYILTELVEP---YTEYQPLWEPKIP-LGRLGRPEELA--G
MMAR_2849|M.marinum_M QIRVNSVSPGYIRTELVEP---LADYHALWEPKIP-LGRMGRPEELT--G
MAV_2775|M.avium_104 DIRVNSVSPGYIRTELVEP---LHEYHRQWQPKIP-LGRMGRPDELA--G
Mflv_0305|M.gilvum_PYR-GCK GIRANTVAPGWFETEMTSEGMADERFTSRVLPRVP-VRRWGAAEDVAGVG
TH_4266|M.thermoresistible__bu GVRVNCIACGAIRTPGLEADAKRQGFDIDMIGQTNGSGRIAEPDEIG--Y
MSMEG_3420|M.smegmatis_MC2_155 GLQINGLAPGYIHTEMTQALVDDTSFNNWILGRTP-AGRWGTVDDLV--G
MLBr_01807|M.leprae_Br4923 SITANVVAPGFIDTEMTRAMDSKYQEAALQ--AIP-LGRIGAVEEIA--G
MAB_2723c|M.abscessus_ATCC_199 SITANVVAPGFIDTDMTRELGEKHVETALT--AIP-MGRIGEPSEVA--G
MUL_1491|M.ulcerans_Agy99 NVTANVVAPGYIDTDMTRALDERIQEGALQ--FIP-AKRVGTAAEVA--G
Mvan_0894|M.vanbaalenii_PYR-1 NVRVNALAPGFFPSELTTELLDDDAGGAYIRRNTP-MARAGRAGELS--G
: * :: * : : * . ::
Mb1963c|M.bovis_AF2122/97 LYLYLASEASSYMTGSDIVIDGGYTCP--
Rv1928c|M.tuberculosis_H37Rv LYLYLASEASSYMTGSDIVIDGGYTCP--
MMAR_2849|M.marinum_M LYLYLASAASSYMTGSDIVIDGGYTCP--
MAV_2775|M.avium_104 IYLYLASAASSYMTGSDIVIDGGYSCP--
Mflv_0305|M.gilvum_PYR-GCK VAVYLAGPASAYHTGDVLRLDGGYLKF--
TH_4266|M.thermoresistible__bu GVLFFASDASSYCSGQTLYMHGGPGPAGV
MSMEG_3420|M.smegmatis_MC2_155 PAIFLASPASAYVNGQVLFVDGGMTAVV-
MLBr_01807|M.leprae_Br4923 AVSFLASEDAGYISGAVIPVDGGLGMGH-
MAB_2723c|M.abscessus_ATCC_199 VVSWLASPDAAYVSGAVIPVDGGLGMGH-
MUL_1491|M.ulcerans_Agy99 VVSFLASEDASYISGAVIPVDGGMGMGH-
Mvan_0894|M.vanbaalenii_PYR-1 ALLLLASGAGSFITGQVIAVDGGWTAR--
:*. ..: .* : :.**