For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MANPTRRTRSDALQNRARILEVAAKAFAEQGLDTAPAAIAKLAGVGIGTLYRHFPTRETLIDAAYRHQLT RVCDAVADLAARHPGDEATRRWMERFIDYATAKSGMSEALNAVIASGADPFADSLSMLTEAVGTLLEAGR RDHTLRTDVTAGDVLLFMGGIAYAAQHGTRDQARRLVDLFIDALTKDTRPDT
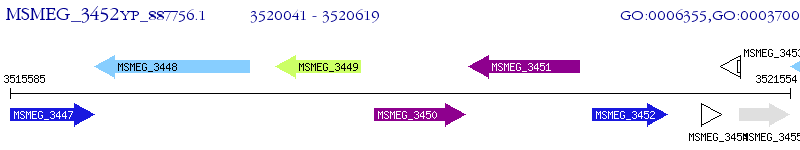
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3452 | - | - | 100% (192) | TetR family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_2305 | - | 2e-23 | 33.51% (185) | TetR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1025 | - | 2e-23 | 33.51% (185) | TetR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0068c | - | 1e-23 | 38.10% (189) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0142 | - | 1e-16 | 35.37% (147) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0067c | - | 1e-23 | 38.10% (189) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00316 | - | 2e-18 | 34.57% (188) | putative TetR/AcrR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1873c | - | 1e-33 | 42.16% (185) | TetR family transcriptional regulator |
| M. marinum M | MMAR_1145 | - | 6e-09 | 30.99% (142) | transcriptional regulatory protein |
| M. avium 104 | MAV_4593 | - | 1e-36 | 44.94% (178) | TetR family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_2070 | - | 2e-06 | 35.90% (78) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_0910 | - | 1e-08 | 30.28% (142) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_0763 | - | 6e-19 | 38.67% (150) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1145|M.marinum_M -----------MAATRTTARAGGDR-RAAIIEAALNRFLSQGIDATALRQ
MUL_0910|M.ulcerans_Agy99 -----------MAATRTTARAGGDR-RAAIIEAALNRFLSQGIDATALRQ
Mb0068c|M.bovis_AF2122/97 -----------MAPTDRRVRADAARNRARVLEVAYQTFAADGLSV-PVDE
Rv0067c|M.tuberculosis_H37Rv -----------MAPTDRRVRADAARNRARVLEVAYQTFAADGLSV-PVDE
MLBr_00316|M.leprae_Br4923 MSVPRSGLSVQMAQPARPLRADAARNRARILGVAYEAFATEGLSV-PIDE
Mflv_0142|M.gilvum_PYR-GCK --------------MTRPLRADAARNRARVLEVAYDTFAAEGLTV-PIDE
Mvan_0763|M.vanbaalenii_PYR-1 --------------MTRPLRADAARNRARVLEVAYDTFAAEGLGV-PIDE
MAB_1873c|M.abscessus_ATCC_199 ----------MSEAELRTPRADAARNRDQLLAAATRAFASADSEP-SMRA
MAV_4593|M.avium_104 ----------------MVGRSDARRNRQRLLEAATAAFTAHGASV-SLES
MSMEG_3452|M.smegmatis_MC2_155 -----------MANPTRRTRSDALQNRARILEVAAKAFAEQGLDT-APAA
TH_2070|M.thermoresistible__bu -----------------MTNSGPPTRRDELLELAAQMFAERGLKATTVRD
.:. * :: * * . .
MMAR_1145|M.marinum_M IQRDAGVSNGSFFHHFPSKEVLAAAVYVDCVTRYQQALLKDLARYPDAES
MUL_0910|M.ulcerans_Agy99 IQRDAGVSNGSFFHHFPSKEVLTAAVYVDCVTRYQQALLKDLVRYPDAES
Mb0068c|M.bovis_AF2122/97 IARRAGVGAGTVYRHFPTKEALFQAVIADRMHRIIDKGHALLKSKHPGDA
Rv0067c|M.tuberculosis_H37Rv IARRAGVGAGTVYRHFPTKEALFQAVIADRMHRIIDKGHALLKSKHPGDA
MLBr_00316|M.leprae_Br4923 IARRAGVGAGTVYRHFPTKEALCAAVIGDRMPHLVDDGYVLLKSAGPGEA
Mflv_0142|M.gilvum_PYR-GCK IARRAGVGAGTVYRHFPTKEALVVAVAEDRVRRIAERARTLLAVEGPGQA
Mvan_0763|M.vanbaalenii_PYR-1 IARRAGVGAGTVYRHFPTKQALVVAVAEDRVRRIVDHARTLLAAEGPGEA
MAB_1873c|M.abscessus_ATCC_199 IARQAGVGIATLYRHFPTRESLVDAVYRDQVVRLTTGAHELLGQ-LPPAT
MAV_4593|M.avium_104 IARDAGVGIGTLYRHFPNREALVEAVYRAELAEVAAAAAQLLQR-HPPKA
MSMEG_3452|M.smegmatis_MC2_155 IAKLAGVGIGTLYRHFPTRETLIDAAYRHQLTRVCDAVADLAAR-HPGDE
TH_2070|M.thermoresistible__bu IADSAGILSGSLYHHFASKEEMVDEVLRGFLDWLFDRYRSIVETEPNPLE
* **: .:.::**..:: : . :
MMAR_1145|M.marinum_M AVRGIVGMHLSWCTEHPEMARFLITMTEPAVHRAAADELRSHNERFATAV
MUL_0910|M.ulcerans_Agy99 AVRGIVGMHLSWCTEHPEMARFLITMTEPAVHRAAADELRSHNERFATAV
Mb0068c|M.bovis_AF2122/97 LFAFLRSMVLQWGATDRGLVEALAGVG-IEISSAAP----EAEADFLDLL
Rv0067c|M.tuberculosis_H37Rv LFAFLRSMVLQWGATDRGLVEALAGVG-IEISSAAP----EAEADFLDLL
MLBr_00316|M.leprae_Br4923 LFTYLRSLVLHWGATDRGLVDALAGAGGIDILGVAP----DAEDAFLTIL
Mflv_0142|M.gilvum_PYR-GCK LFVFFRDMVRS-AAVDHGLVDALMNQG-LDLEAVAP----GVEAAFLSTL
Mvan_0763|M.vanbaalenii_PYR-1 LFVFLRDMVRS-AAADYGLVDALVGYG-LDLEVAAP----GAEAAFLATL
MAB_1873c|M.abscessus_ATCC_199 AMRRWMDSFGDWIATKNGMLDTLHTMIESGHIGHA-----ETRAELLKAV
MAV_4593|M.avium_104 ALRRWMDRYATFVATKRGMAESLQAIFESGALVPS-----QTRDSIVGAV
MSMEG_3452|M.smegmatis_MC2_155 ATRRWMERFIDYATAKSGMSEALNAVIASGADPFA-----DSLSMLTEAV
TH_2070|M.thermoresistible__bu RFKGLFMASFEAIEHRHAQVVIYQDEAKRLSSQPRFAYVEQRNREQRKMW
MMAR_1145|M.marinum_M QTWWRPHAHYGALRPLSPAHSQALWLGPAQELVRAWLLGNLAAPPNAAD-
MUL_0910|M.ulcerans_Agy99 QTWWRPHAHYGALRPLSPAHSQALWLGPAQELVRAWLLGNLAAPPNAAD-
Mb0068c|M.bovis_AF2122/97 TDLLRAAQRAGTVR-----------PDVDVLEVKTLLVGCQAMQSYNAE-
Rv0067c|M.tuberculosis_H37Rv TDLLRAAQRAGTVR-----------PDVDVLEVKTLLVGCQAMQSYNAE-
MLBr_00316|M.leprae_Br4923 SDLLYAAQYAGTAR-----------TDVGVREVKSILVGCQAMEAYNSA-
Mflv_0142|M.gilvum_PYR-GCK GDLLSAAQGAGTVR-----------PDVDVAVVKALLVVCKVPQVFDSE-
Mvan_0763|M.vanbaalenii_PYR-1 GELLAAAQRAGTVR-----------ADVDVAVIKALLVVCKVPQVYDGE-
MAB_1873c|M.abscessus_ATCC_199 TTILDAGHAAGDLR-----------ADVTAEDIAASLIGIFTVAPRPEQE
MAV_4593|M.avium_104 ETLLRAGADDASLR-----------ADVQADDVVSSLIGIFLVSGSPEQ-
MSMEG_3452|M.smegmatis_MC2_155 GTLLEAGRRDHTLR-----------TDVTAGDVLLFMGGIAYAAQHGTR-
TH_2070|M.thermoresistible__bu VDVLTQGIEEGYFR-----------SDIDVDLVYRFIRDTTWVSVRWYRP
* . : :
MMAR_1145|M.marinum_M ---------AAILADAAWLCLRAGPAG---
MUL_0910|M.ulcerans_Agy99 ---------AAILADAAWLCLRAGPAG---
Mb0068c|M.bovis_AF2122/97 --------LAAKVTDVALDGLRANRK----
Rv0067c|M.tuberculosis_H37Rv --------LAAKVTDVALDGLRANRK----
MLBr_00316|M.leprae_Br4923 --------LAERVTDVVVDGLRAAR-----
Mflv_0142|M.gilvum_PYR-GCK --------VTERVTRVIEDGLRPLR-----
Mvan_0763|M.vanbaalenii_PYR-1 --------VSERVVRVIEDGLRVRSSV---
MAB_1873c|M.abscessus_ATCC_199 -------AQADRLLNLLMDGLRPAARQAHS
MAV_4593|M.avium_104 ---------TGRMLDLLVAGVTR-------
MSMEG_3452|M.smegmatis_MC2_155 -------DQARRLVDLFIDALTKDTRPDT-
TH_2070|M.thermoresistible__bu GGPLTAEEVGRQYLSIVLGGITAGAN----
: