For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRTMGGHQTQITQLAGATPGLLARELETPSRVDEITDRLVTAIAIGEYLPGARLPSERELASSLRVGRMT VRAALARLVDLGLVETKSAGRGGGTYVLQQWPESSTEAVGRTLTMRVDELRDRCDAICRIHGAVCRAAAE TASEQDVGLLRERLEAYRAAPSGLQAQQADSALHLAIMDASGNAVLKQVLLDLEASVSIGAPAHLWGEPG TMRDMELRALREHEQLIDAIARGDGDEADDLARAHVAIDFELISAAMRRAGVLAE
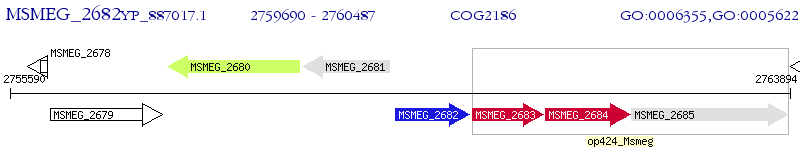
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2682 | - | - | 100% (265) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6700 | - | 7e-20 | 35.19% (216) | regulatory protein |
| M. smegmatis MC2 155 | MSMEG_1117 | - | 1e-18 | 32.89% (225) | transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0505 | - | 9e-11 | 32.50% (160) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4061 | - | 2e-16 | 30.84% (227) | regulatory protein GntR, HTH |
| M. tuberculosis H37Rv | Rv0494 | - | 9e-11 | 32.50% (160) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4644c | - | 1e-10 | 27.68% (224) | GntR family transcriptional regulator |
| M. marinum M | MMAR_0749 | - | 3e-15 | 30.63% (222) | GntR family transcriptional regulator |
| M. avium 104 | MAV_2560 | - | 2e-11 | 25.53% (235) | regulatory protein GntR, HTH |
| M. thermoresistible (build 8) | TH_4115 | - | 1e-11 | 30.80% (224) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_1381 | - | 1e-14 | 30.63% (222) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1846 | - | 3e-19 | 34.26% (216) | regulatory protein GntR, HTH |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0749|M.marinum_M ----------------------MQTVRRRTLIAQVSDQLRDEITSGRWKV
MUL_1381|M.ulcerans_Agy99 -------------------------MRRRTLIAQVSDQLRDEITSGRWKV
MAB_4644c|M.abscessus_ATCC_199 ---------------------MHPRLSSQSLAEQVANILAAQIGDGRWST
Mb0505|M.bovis_AF2122/97 ------------MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPP
Rv0494|M.tuberculosis_H37Rv ------------MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPP
TH_4115|M.thermoresistible__bu ---------------------------VQSSYIQVADQLRDLILRGDLAV
MAV_2560|M.avium_104 --------------------MEVTSLREPKMADRVATVLRRMFIRGEITE
Mflv_4061|M.gilvum_PYR-GCK ------MAAPDPADSSDASEALLRPVRPLNAFEDTVERLLQTIRLGVLTP
MSMEG_2682|M.smegmatis_MC2_155 MRTMGGHQTQITQLAGATPGLLARELETPSRVDEITDRLVTAIAIGEYLP
Mvan_1846|M.vanbaalenii_PYR-1 ---------------MSSLSPLVAAEAPVGRADEIVQRITEAIHLGLLDD
: : *
MMAR_0749|M.marinum_M GDRIPTEPELCEMTGTARNTVREAVQALVHVGILERRQGS--GTYVIA--
MUL_1381|M.ulcerans_Agy99 GDRIPTEPELCEMTGTARNTVREAVQALVHVGILERRQGS--GTYVIA--
MAB_4644c|M.abscessus_ATCC_199 GAKLPGEVALCEEFGVSRATVREAIRDLKTKGLVSTRQGA--GVFVTS--
Mb0505|M.bovis_AF2122/97 GSTLPPERDLAERLGVNRTSLRQGLARLQQMGLIEVRHGS--GSVVRDPE
Rv0494|M.tuberculosis_H37Rv GSTLPPERDLAERLGVNRTSLRQGLARLQQMGLIEVRHGS--GSVVRDPE
TH_4115|M.thermoresistible__bu GQKLPSEAEMAPLLGVSRSTIREALRILTTEHLIETRRGIQGGAFVAVPD
MAV_2560|M.avium_104 GTMLPPESELMERFGVSRPTLREAFRVLESESLIQVQRGVRGGARVTRPR
Mflv_4061|M.gilvum_PYR-GCK GESLPPERELAARLGVSRDTVREAIKSLADAGYLVSRRGRYGGTFLAD--
MSMEG_2682|M.smegmatis_MC2_155 GARLPSERELASSLRVGRMTVRAALARLVDLGLVETKSAGRGGGTYVLQQ
Mvan_1846|M.vanbaalenii_PYR-1 GERLPVEVDLAAQFGVAPMTVREALATLREQELVETRRGRSGGSFVRRPA
* :* * : . ::* .. * : : . *
MMAR_0749|M.marinum_M ---DVEQGSALAEYFAAARERDLWELR------EAVEVTAAVLSARRRDA
MUL_1381|M.ulcerans_Agy99 ---DVEQGSALAEYFAAARERDLWELR------EAVEVTAAVLSARRRDA
MAB_4644c|M.abscessus_ATCC_199 ---DRPLG-QWDRVLAAAEIDAVVEIR------RLLEVQAAGLAAERRNR
Mb0505|M.bovis_AF2122/97 GLTHPAVVEALVRKLGPDFLVELLEIR------AALGPLIGRLAAARSTP
Rv0494|M.tuberculosis_H37Rv GLTHPAVVEALVRKLGPDFLVELLEIR------AALGPLIGRLAAARSTP
TH_4115|M.thermoresistible__bu ---SERLEGMFNTSFSMLALTNQVDARSFLQAMRALEGPAAALAAAARGG
MAV_2560|M.avium_104 R-ETLARYAGLILEYEGVTVKDVYDAR------VTLEVPMVEQLAKDRNP
Mflv_4061|M.gilvum_PYR-GCK ---ALPAPRADGTEMHRDDIDDAVRLR------EILEVGAARMAAVRTLA
MSMEG_2682|M.smegmatis_MC2_155 WPESSTEAVGRTLTMRVDELRDRCDAI------CRIHGAVCRAAAETASE
Mvan_1846|M.vanbaalenii_PYR-1 G-PPVDKLTARLAAMSASDLRDLFDEH------TAVSGQAARLAAERAAP
: *
MMAR_0749|M.marinum_M ADIEDLRALLARRNELWSREPANDKERTEMTSADTALHRAIVAASHNEIY
MUL_1381|M.ulcerans_Agy99 ADIEDLRVLLARRNELWSREPANDKERTEMTSADTALHRAIVAASHNETY
MAB_4644c|M.abscessus_ATCC_199 SDVAAIRRALVAR-----AKAESAGDDIEYVDADLEFHRAVVAAVHNPLL
Mb0505|M.bovis_AF2122/97 EDAEALCAALEVV--------QQADTAAARQAADLAYFRVLIHSTRNRAL
Rv0494|M.tuberculosis_H37Rv EDAEALCAALEVV--------QQADTAAARQAADLAYFRVLIHSTRNRAL
TH_4115|M.thermoresistible__bu DQVEELKRTAAMV---------DPADQEAAMQQSADFHSAILRASGNKLL
MAV_2560|M.avium_104 KVIAELEQIVERE--------SQLNPGSEAVDQLTDFHAAIARLSGNSTL
Mflv_4061|M.gilvum_PYR-GCK AVEREALRSRLTD--------VRGADPADYRRLDSRLHLAIAEAAGVPSL
MSMEG_2682|M.smegmatis_MC2_155 QDVGLLRERLEAY--------RAAPSGLQAQQADSALHLAIMDASGNAVL
Mvan_1846|M.vanbaalenii_PYR-1 YAVRRLFALTDQLG--------AAATLGDRIRADSRFHIQVAMASQSARL
. :
MMAR_0749|M.marinum_M LQFYDLLLPTIHRAVEAGPVGT-----CTS--YEREHTELVEAIIAGDPQ
MUL_1381|M.ulcerans_Agy99 LQFYDLLLPTIHRAVEAGPVGT-----CTS--YEREHTELVEAIIAGDPQ
MAB_4644c|M.abscessus_ATCC_199 DELFDSFSVRMREAMLALLAGS-----AERPHDQEHHQWLFDAIVDGSTR
Mb0505|M.bovis_AF2122/97 GLLYRWVEHAFGGREHALTGAY-----DDADPVLTDLRAINGAVLAGDPA
Rv0494|M.tuberculosis_H37Rv GLLYRWVEHAFGGREHALTGAY-----DDADPVLTDLRAINGAVLAGDPA
TH_4115|M.thermoresistible__bu EAMGRPVSAVARSKFRRTVPGL-----EFWERNQRAHQELLKAILAGDEN
MAV_2560|M.avium_104 QIVSDMLHHIIEKANRSLQPTKGTRAEQAVRRSAKTHRMVLDMIKEGDAE
Mflv_4061|M.gilvum_PYR-GCK VPLVAQNRMRLNELLDRIPLMT-----RNIAHSDEQHEAIVSAILAGDAD
MSMEG_2682|M.smegmatis_MC2_155 KQVLLDLEASVSIGAPAHLWGEPGTMRDMELRALREHEQLIDAIARGDGD
Mvan_1846|M.vanbaalenii_PYR-1 ARREANLQAEAAGLVWLPIGPS-----LDVAAHVEEQHAIAAAVASEMAD
: :
MMAR_0749|M.marinum_M RAEAAAQVFIARLRP---------------------
MUL_1381|M.ulcerans_Agy99 RAEAAAQVFIARLRR---------------------
MAB_4644c|M.abscessus_ATCC_199 RAVELATDQLQHVRMR--------------------
Mb0505|M.bovis_AF2122/97 AAAATVEAYLNASALRMVKSYRDRA-----------
Rv0494|M.tuberculosis_H37Rv AAAATVEAYLNASALRMVKSYRDRA-----------
TH_4115|M.thermoresistible__bu LAYELATRHIDEDLSPYYLPPEEADRDGAGA-----
MAV_2560|M.avium_104 KAGELWKRHLQKAEEFVLTGAELSTVVDLLE-----
Mflv_4061|M.gilvum_PYR-GCK TAAQTMAAHVWGSAALLHGFLD--------------
MSMEG_2682|M.smegmatis_MC2_155 EADDLARAHVAIDFELISAAMRRAGVLAE-------
Mvan_1846|M.vanbaalenii_PYR-1 EARRLAEAHVMGQLARVTSINLALSTGELSASGSAS
* :