For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HPRLSSQSLAEQVANILAAQIGDGRWSTGAKLPGEVALCEEFGVSRATVREAIRDLKTKGLVSTRQGAGV FVTSDRPLGQWDRVLAAAEIDAVVEIRRLLEVQAAGLAAERRNRSDVAAIRRALVARAKAESAGDDIEYV DADLEFHRAVVAAVHNPLLDELFDSFSVRMREAMLALLAGSAERPHDQEHHQWLFDAIVDGSTRRAVELA TDQLQHVRMR*
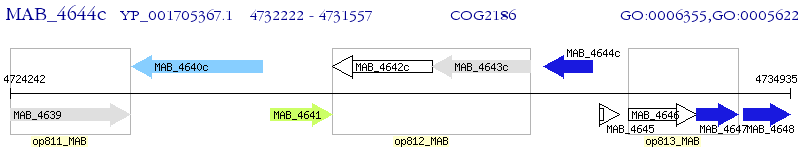
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4644c | - | - | 100% (221) | GntR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2272c | - | 2e-27 | 38.50% (213) | putative transcriptional regulator, GntR |
| M. abscessus ATCC 19977 | MAB_3018 | - | 2e-14 | 28.96% (221) | GntR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0601 | - | 2e-11 | 29.33% (225) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0016 | - | 2e-29 | 38.28% (209) | GntR domain-containing protein |
| M. tuberculosis H37Rv | Rv0586 | - | 2e-11 | 29.33% (225) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0749 | - | 4e-24 | 35.44% (206) | GntR family transcriptional regulator |
| M. avium 104 | MAV_2560 | - | 2e-12 | 28.57% (224) | regulatory protein GntR, HTH |
| M. smegmatis MC2 155 | MSMEG_2794 | - | 6e-29 | 37.37% (198) | transcriptional regulator, GntR family protein |
| M. thermoresistible (build 8) | TH_4115 | - | 2e-17 | 33.94% (218) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_1381 | - | 9e-24 | 35.44% (206) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0952 | - | 5e-26 | 33.49% (209) | GntR domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4644c|M.abscessus_ATCC_199 ---------MHPRLSSQSLAEQVANILAAQIGDGRWSTGAKLPGEVALCE
Mflv_0016|M.gilvum_PYR-GCK MIGRKDIRVPLSTARRSGLVEQVIDQLRHAVAGQEWRIGDRIPNETVLVE
Mvan_0952|M.vanbaalenii_PYR-1 --------MPLSTARRTGLVDQVIEQLRRAVAGGEWRVGERIPNETVLVE
MSMEG_2794|M.smegmatis_MC2_155 --------MPLASTRRTGLVDQVIEQLRASVSSGEWPVGTKIPTEPELAD
MMAR_0749|M.marinum_M ----------MQTVRRRTLIAQVSDQLRDEITSGRWKVGDRIPTEPELCE
MUL_1381|M.ulcerans_Agy99 -------------MRRRTLIAQVSDQLRDEITSGRWKVGDRIPTEPELCE
Mb0601|M.bovis_AF2122/97 --------MALQPVTRRSVPEEVFEQIATDVLTGEMPPGEALPSERRLAE
Rv0586|M.tuberculosis_H37Rv --------MALQPVTRRSVPEEVFEQIATDVLTGEMPPGEALPSERRLAE
TH_4115|M.thermoresistible__bu ---------------VQSSYIQVADQLRDLILRGDLAVGQKLPSEAEMAP
MAV_2560|M.avium_104 --------MEVTSLREPKMADRVATVLRRMFIRGEITEGTMLPPESELME
.* : . * :* * :
MAB_4644c|M.abscessus_ATCC_199 EFGVSRATVREAIRDLKTKGLVSTRQ--GAGVFVTSDR----PLG-QWDR
Mflv_0016|M.gilvum_PYR-GCK KFGVGRNTVREAVRALAHAGILEVRQ--GDGTYVRATS----EVSGALRR
Mvan_0952|M.vanbaalenii_PYR-1 SLGVGRNTVREAVRALAHAGILEVRQ--GDGTYVRATS----EVSGALRR
MSMEG_2794|M.smegmatis_MC2_155 TLGVGRNTLREAVRALAHSGILEVRQ--GDGTYVRATS----EVSGALRR
MMAR_0749|M.marinum_M MTGTARNTVREAVQALVHVGILERRQ--GSGTYVIADV----EQGSALAE
MUL_1381|M.ulcerans_Agy99 MTGTARNTVREAVQALVHVGILERRQ--GSGTYVIADV----EQGSALAE
Mb0601|M.bovis_AF2122/97 LLGVSRPAVREALKRLSAAGLVEVRQ--GDVTTVRDFRR--HAGLDLLPR
Rv0586|M.tuberculosis_H37Rv LLGVSRPAVREALKRLSAAGLVEVRQ--GDVTTVRDFRR--HAGLDLLPR
TH_4115|M.thermoresistible__bu LLGVSRSTIREALRILTTEHLIETRRGIQGGAFVAVPDS--ERLEGMFNT
MAV_2560|M.avium_104 RFGVSRPTLREAFRVLESESLIQVQRGVRGGARVTRPRRETLARYAGLIL
*..* ::***.: * ::. :: . *
MAB_4644c|M.abscessus_ATCC_199 VLAAAEIDAVVEIRRLLEVQAAGLAAERRNRSDVAAIRRALVAR-----A
Mflv_0016|M.gilvum_PYR-GCK -LCGEELREVLEVRRALEVEAARLAASRRTPADVAEMRGLLARRD---ER
Mvan_0952|M.vanbaalenii_PYR-1 -LCGAELREVLQVRRALEVEGARLAAAHCTPADIAEMRTLLARRD---EC
MSMEG_2794|M.smegmatis_MC2_155 -LCGSELREVLQVRRCLEVEGARLAASNRTDADLAELRAFLDR-------
MMAR_0749|M.marinum_M YFAAARERDLWELREAVEVTAAVLSARRRDAADIEDLRALLARRNELWSR
MUL_1381|M.ulcerans_Agy99 YFAAARERDLWELREAVEVTAAVLSARRRDAADIEDLRVLLARRNELWSR
Mb0601|M.bovis_AF2122/97 LLFRNGELDISVVRSILEARLRNFPKVAELAAERNEPELAELLQDS--LR
Rv0586|M.tuberculosis_H37Rv LLFRNGELDISVVRSILEARLRNFPKVAELAAERNEPELAELLQDS--LR
TH_4115|M.thermoresistible__bu SFSMLALTNQVDARSFLQAMRALEGPAAALAAAARGGDQVEELKR---TA
MAV_2560|M.avium_104 EYEGVTVKDVYDARVTLEVPMVEQLAKDRNPKVIAELEQIVERES-----
* ::.
MAB_4644c|M.abscessus_ATCC_199 KAESAGDDIEYVDADLEFHRAVVAAVHNPLLDELFDSFSVRMREAMLALL
Mflv_0016|M.gilvum_PYR-GCK DSAGDVE--AFARIDTEFHLAVVRCARNNTLIELYRGLLEAVTASVAH--
Mvan_0952|M.vanbaalenii_PYR-1 AATDDTD--AFARTDAEFHLAVVRGSGNRTLTELYRGLIEVVTASVAT--
MSMEG_2794|M.smegmatis_MC2_155 TETSDDD--DFVHSDTAFHLAVVRASHNSVLIELYRGLIEAITASVAT--
MMAR_0749|M.marinum_M EPANDKERTEMTSADTALHRAIVAASHNEIYLQFYDLLLPTIHRAVEAGP
MUL_1381|M.ulcerans_Agy99 EPANDKERTEMTSADTALHRAIVAASHNETYLQFYDLLLPTIHRAVEAGP
Mb0601|M.bovis_AF2122/97 ALDTEEDPIVWQRHTLDFWDHVVDSAGSIVDRLMYNAFRAAYEPTLAALT
Rv0586|M.tuberculosis_H37Rv ALDTEEDPIVWQRHTLDFWDHVVDSAGSIVDRLMYNAFRAAYEPTLAALT
TH_4115|M.thermoresistible__bu AMVDPADQEAAMQQSADFHSAILRASGNKLLEAMGRPVSAVARSKFRRTV
MAV_2560|M.avium_104 ---QLNPGSEAVDQLTDFHAAIARLSGNSTLQIVSDMLHHIIEKANRSLQ
: : . . .
MAB_4644c|M.abscessus_ATCC_199 AGSAERPHDQEH-----HQWLFDAIVDGSTRRAVELATDQLQHVRMR---
Mflv_0016|M.gilvum_PYR-GCK TRHVP--VDVCD-----HGVLLDHIAAGDPDLAARTAGEFLDRIINGLGS
Mvan_0952|M.vanbaalenii_PYR-1 TRHVP--VEMVD-----HGVLVDQIAAGDAEQAARTAGELLDRILAGVDS
MSMEG_2794|M.smegmatis_MC2_155 ASARP--DGKVS-----HRGLVEAIAAGDVERAGREAGGFLDDLLAQTPD
MMAR_0749|M.marinum_M VGTCT--SYERE-----HTELVEAIIAGDPQRAEAAAQVFIARLRP----
MUL_1381|M.ulcerans_Agy99 VGTCT--SYERE-----HTELVEAIIAGDPQRAEAAAQVFIARLRR----
Mb0601|M.bovis_AF2122/97 TTMTAAAKRPSD-----YRKLADAICSGDPTGAKKAAQDLLELANTSLMA
Rv0586|M.tuberculosis_H37Rv TTMTAAAKRPSD-----YRKLADAICSGDPTGAKKAAQDLLELANTSLMA
TH_4115|M.thermoresistible__bu PGLEFWERNQRA-----HQELLKAILAGDENLAYELATRHIDEDLSPYYL
MAV_2560|M.avium_104 PTKGTRAEQAVRRSAKTHRMVLDMIKEGDAEKAGELWKRHLQKAEEFVLT
: : . * *. * :
MAB_4644c|M.abscessus_ATCC_199 ------------
Mflv_0016|M.gilvum_PYR-GCK ADSS--------
Mvan_0952|M.vanbaalenii_PYR-1 ADSC--------
MSMEG_2794|M.smegmatis_MC2_155 PQG---------
MMAR_0749|M.marinum_M ------------
MUL_1381|M.ulcerans_Agy99 ------------
Mb0601|M.bovis_AF2122/97 VLVSQASRQ---
Rv0586|M.tuberculosis_H37Rv VLVSQASRQ---
TH_4115|M.thermoresistible__bu PPEEADRDGAGA
MAV_2560|M.avium_104 GAELSTVVDLLE