For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AAPDPADSSDASEALLRPVRPLNAFEDTVERLLQTIRLGVLTPGESLPPERELAARLGVSRDTVREAIKS LADAGYLVSRRGRYGGTFLADALPAPRADGTEMHRDDIDDAVRLREILEVGAARMAAVRTLAAVEREALR SRLTDVRGADPADYRRLDSRLHLAIAEAAGVPSLVPLVAQNRMRLNELLDRIPLMTRNIAHSDEQHEAIV SAILAGDADTAAQTMAAHVWGSAALLHGFLD*
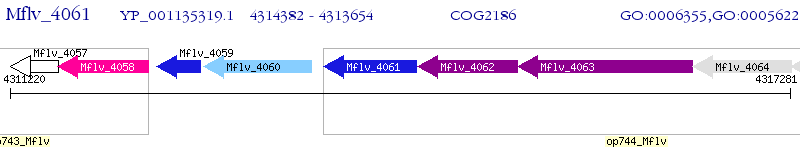
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4061 | - | - | 100% (242) | regulatory protein GntR, HTH |
| M. gilvum PYR-GCK | Mflv_0016 | - | 3e-14 | 29.72% (212) | GntR domain-containing protein |
| M. gilvum PYR-GCK | Mflv_0958 | - | 1e-09 | 31.16% (215) | GntR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0601 | - | 2e-10 | 29.57% (230) | GntR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0586 | - | 2e-10 | 29.57% (230) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4644c | - | 2e-14 | 30.85% (201) | GntR family transcriptional regulator |
| M. marinum M | MMAR_0820 | - | 3e-12 | 29.34% (242) | GntR family transcriptional regulator |
| M. avium 104 | MAV_3932 | - | 3e-09 | 31.71% (205) | transcriptional regulator, GntR family protein |
| M. smegmatis MC2 155 | MSMEG_2599 | - | 1e-97 | 77.69% (242) | GntR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_4115 | - | 6e-14 | 28.77% (219) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_4564 | - | 4e-12 | 28.93% (242) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2282 | - | 1e-109 | 84.77% (243) | regulatory protein GntR, HTH |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4061|M.gilvum_PYR-GCK -MAAPDPADS------------------------------SD--------
Mvan_2282|M.vanbaalenii_PYR-1 MSAVPEPTDERLREEQTASDERLREEQTASDERLREEQTASDGADATAEQ
MSMEG_2599|M.smegmatis_MC2_155 MSADPDPQPT------------------------------AE--------
Mb0601|M.bovis_AF2122/97 --------------------------------------------------
Rv0586|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0820|M.marinum_M --------------------------------------------------
MUL_4564|M.ulcerans_Agy99 --------------------------------------------------
MAB_4644c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4115|M.thermoresistible__bu --------------------------------------------------
MAV_3932|M.avium_104 --------------------------------------------------
Mflv_4061|M.gilvum_PYR-GCK -ASEALLRPVRPLNAFEDTVERLLQTIRLGVLTPGESLPPERELAARLGV
Mvan_2282|M.vanbaalenii_PYR-1 IAADALLRPVRPLNAFEDTVERLLQTIRLGVLRPGESLPPERELATRLGV
MSMEG_2599|M.smegmatis_MC2_155 -AADALLRPVRLGNAFEDTVGRLLETIRLGVLGPGESLPPERELATRLGV
Mb0601|M.bovis_AF2122/97 ----MALQPVTRRSVPEEVFEQIATDVLTGEMPPGEALPSERRLAELLGV
Rv0586|M.tuberculosis_H37Rv ----MALQPVTRRSVPEEVFEQIATDVLTGEMPPGEALPSERRLAELLGV
MMAR_0820|M.marinum_M MNQSSILQPPDRQRVDEQIATSIADAILDGVFPPGSALPPERELAEQLGV
MUL_4564|M.ulcerans_Agy99 MNQSSILQPPDRQRVDEQIATSIADAILDGVFPPGSALPPERELAEQLGV
MAB_4644c|M.abscessus_ATCC_199 -----MHPRLSSQSLAEQVANILAAQIGDGRWSTGAKLPGEVALCEEFGV
TH_4115|M.thermoresistible__bu -----------VQSSYIQVADQLRDLILRGDLAVGQKLPSEAEMAPLLGV
MAV_3932|M.avium_104 --MNAEPRTGPAKTLASALARDIEAEIVRRGWAVGESLGSEPALQQRFGV
: : * * * : :**
Mflv_4061|M.gilvum_PYR-GCK SRDTVREAIKSLADAGYLVSRRGRYG--GTFLADALPAPRADGTEMHRDD
Mvan_2282|M.vanbaalenii_PYR-1 SRDTVREAIKSLADAGYLVSRRGRYG--GTFIADALPAPRTGDVAFSRTD
MSMEG_2599|M.smegmatis_MC2_155 SRDTVREAIKSLADAGYLVSRRGRYG--GTFLADELPSHTADTPRPTHEE
Mb0601|M.bovis_AF2122/97 SRPAVREALKRLSAAGLVEVRQGDVT--TVRDFRRHAGLDLLPRLLFRNG
Rv0586|M.tuberculosis_H37Rv SRPAVREALKRLSAAGLVEVRQGDVT--TVRDFRRHAGLDLLPRLLFRNG
MMAR_0820|M.marinum_M NRTSLRQGLARLQQMGLIETRQGSGN--VVRDPQSLTHPAVVEALVRKLG
MUL_4564|M.ulcerans_Agy99 NRTSLRQGLARLQQMGLIETRQGSGN--VVRDPQSLTHPAVVEALVRKLG
MAB_4644c|M.abscessus_ATCC_199 SRATVREAIRDLKTKGLVSTRQGAGV--FVTSDRPLGQWDRVLAAAEIDA
TH_4115|M.thermoresistible__bu SRSTIREALRILTTEHLIETRRGIQGGAFVAVPDSERLEGMFNTSFSMLA
MAV_3932|M.avium_104 SRSVLREAVRLVEHHQVARMRRGPNGGLYICEPDAGPATRAVVIYLEYLG
.* :*:.: : *:*
Mflv_4061|M.gilvum_PYR-GCK IDDAVRLREILEVGAARMAAVRTLAAVER-----EALRSRLTDVRGAD-P
Mvan_2282|M.vanbaalenii_PYR-1 IDDALRLREILETGAARMAACRTLSATER-----EALWSRLLDVRGAA-P
MSMEG_2599|M.smegmatis_MC2_155 IDDALRLREILEVGAARMAAGRTLSAAER-----DGLWSRLADVRAAEGT
Mb0601|M.bovis_AF2122/97 ELDISVVRSILEARLRNFPKVAELAAERNEPELAELLQDSLRALDTEEDP
Rv0586|M.tuberculosis_H37Rv ELDISVVRSILEARLRNFPKVAELAAERNEPELAELLQDSLRALDTEEDP
MMAR_0820|M.marinum_M ---PDFLVEVLEIRAALGPLIGRLAAERNSSEDAEALRAALAEVADADTA
MUL_4564|M.ulcerans_Agy99 ---PDFLVEVLEIRAALGPLIGRLAAERNSSEDAEALRAALAEVADADTA
MAB_4644c|M.abscessus_ATCC_199 VVEIRRLLEVQAAGLAAERRNRSDVAAIR------RALVARAKAESAGDD
TH_4115|M.thermoresistible__bu LTNQVDARSFLQAMRALEGPAAALAAAARGGDQVEELKRTAAMVDPADQE
MAV_3932|M.avium_104 ----TTLADLLNARLVLEPLAASLAAERIDEAGIARLRAVLHAEQQWRPG
.. *
Mflv_4061|M.gilvum_PYR-GCK ADYRRLDSRLHLAIAEAAGVPSLVPLVAQNR-----MRLNELLDRIPLMT
Mvan_2282|M.vanbaalenii_PYR-1 QDYRRLDSRLHLAITEAAGVPSLVPLVAQNR-----MRLNEMLDCIPLLP
MSMEG_2599|M.smegmatis_MC2_155 DDYRRLDSRLHLAIAEAAGSPSLVPLIAENR-----MRINGLLDRIPLLP
Mb0601|M.bovis_AF2122/97 IVWQRHTLDFWDHVVDSAGSIVDRLMYNAFR-----AAYEPTLAALTTTM
Rv0586|M.tuberculosis_H37Rv IVWQRHTLDFWDHVVDSAGSIVDRLMYNAFR-----AAYEPTLAALTTTM
MMAR_0820|M.marinum_M PARQAADLAYFRVLIHGTRNRALGLLYRWVE-----QAFGGREHELTGAY
MUL_4564|M.ulcerans_Agy99 PARQAADLAYFRVLIHGTRNRALGLLYRWVE-----QAFGGREHDLTGAY
MAB_4644c|M.abscessus_ATCC_199 IEYVDADLEFHRAVVAAVHNPLLDELFDSFS-----VRMREAMLALLAGS
TH_4115|M.thermoresistible__bu AAMQQS-ADFHSAILRASGNKLLEAMGRPVS-----AVARSKFRRTVPGL
MAV_3932|M.avium_104 LPMPRD--EFHIALAEQSKNPVLQLFIKVLMRLTTRYALQSRTDSETEAL
: :
Mflv_4061|M.gilvum_PYR-GCK RNIAHSDEQHEAIVSAILAGDADTAAQTMAAHVWGSAALLHGFLD-----
Mvan_2282|M.vanbaalenii_PYR-1 RNIAHSDEQHEAIVMAILAGDADGAAQTMAAHVWGSAALLHGFLD-----
MSMEG_2599|M.smegmatis_MC2_155 RNIAHSDDQHEAIVMAILAGDADRAATAMLDHVSGSAALLHGFLD-----
Mb0601|M.bovis_AF2122/97 TAAAKRPSDYRKLADAICSGDPTGAKKAAQDLLELANTSLMAVLVSQASR
Rv0586|M.tuberculosis_H37Rv TAAAKRPSDYRKLADAICSGDPTGAKKAAQDLLELANTSLMAVLVSQASR
MMAR_0820|M.marinum_M EDADPVIADLTAINDAVLAGDPETAAAAVEGYLNASALRMIMCYRAAYAG
MUL_4564|M.ulcerans_Agy99 EDADPVIADLTAINDAVLAGDPETAAAAVEGYLNASALRMIMCYRAAYAG
MAB_4644c|M.abscessus_ATCC_199 AERPHDQEHHQWLFDAIVDGSTRRAVELATDQLQHVRMR-----------
TH_4115|M.thermoresistible__bu EFWERNQRAHQELLKAILAGDENLAYELATRHIDEDLSPYYLPPEEADRD
MAV_3932|M.avium_104 EAVDHLHTHHSRIVAAVTAGDPARAKTLSERHVEAVTAWLQRHHAGDRNR
: *: *. * :
Mflv_4061|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2282|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2599|M.smegmatis_MC2_155 --------------------------------------------------
Mb0601|M.bovis_AF2122/97 Q-------------------------------------------------
Rv0586|M.tuberculosis_H37Rv Q-------------------------------------------------
MMAR_0820|M.marinum_M EDPER---------------------------------------------
MUL_4564|M.ulcerans_Agy99 EDPER---------------------------------------------
MAB_4644c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4115|M.thermoresistible__bu GAGA----------------------------------------------
MAV_3932|M.avium_104 GRTPRRPLNSEVPQGKLAEMLAATIGDDIAADGWRVGSVFGTETALLQRY
Mflv_4061|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2282|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2599|M.smegmatis_MC2_155 --------------------------------------------------
Mb0601|M.bovis_AF2122/97 --------------------------------------------------
Rv0586|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0820|M.marinum_M --------------------------------------------------
MUL_4564|M.ulcerans_Agy99 --------------------------------------------------
MAB_4644c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4115|M.thermoresistible__bu --------------------------------------------------
MAV_3932|M.avium_104 RVSRAVFREAVRLLEYHSIAHMRRGPGGGLVIAEPAAQASIDTIALYLQY
Mflv_4061|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2282|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2599|M.smegmatis_MC2_155 --------------------------------------------------
Mb0601|M.bovis_AF2122/97 --------------------------------------------------
Rv0586|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0820|M.marinum_M --------------------------------------------------
MUL_4564|M.ulcerans_Agy99 --------------------------------------------------
MAB_4644c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4115|M.thermoresistible__bu --------------------------------------------------
MAV_3932|M.avium_104 RDPSREDLRCVRDAIEIDNVAKVVKRLAEPQVAAFVASRRSGLPDDSRQT
Mflv_4061|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2282|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2599|M.smegmatis_MC2_155 --------------------------------------------------
Mb0601|M.bovis_AF2122/97 --------------------------------------------------
Rv0586|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0820|M.marinum_M --------------------------------------------------
MUL_4564|M.ulcerans_Agy99 --------------------------------------------------
MAB_4644c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4115|M.thermoresistible__bu --------------------------------------------------
MAV_3932|M.avium_104 PDDVRRAIAEEFDFHVGLAQLAGNAPLDLFLRIIVELFRRHWSSTGQALP
Mflv_4061|M.gilvum_PYR-GCK -------------------------------------------
Mvan_2282|M.vanbaalenii_PYR-1 -------------------------------------------
MSMEG_2599|M.smegmatis_MC2_155 -------------------------------------------
Mb0601|M.bovis_AF2122/97 -------------------------------------------
Rv0586|M.tuberculosis_H37Rv -------------------------------------------
MMAR_0820|M.marinum_M -------------------------------------------
MUL_4564|M.ulcerans_Agy99 -------------------------------------------
MAB_4644c|M.abscessus_ATCC_199 -------------------------------------------
TH_4115|M.thermoresistible__bu -------------------------------------------
MAV_3932|M.avium_104 TWSDVRAVHHAHLRIADAVAAGDLSVASYRLRRHLDAAASWWL