For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKGTQTMHDGPTAMYINGEWVAEGENGTTTLTAPFDGRELAVIPVGGRRDVERAVQAAHNARRTGLPLDR RLTILEAVAASIAARSEDFARAIALECAKPIKTARAEVLRAIDTIKFSVAAARTLGSESVPLDATAAGAG KVTFTKRVPVGVVAAIAPFNFPLNLVVHKVAPALAVGCPVVLKPATQTPLSALLLAEVFHEAGLPAGWLN VVCGSGAEVGGALATHPDVAYVTFTGSPQVGWGIAEAAPKKKVRLELGSNAPLIVDRESDWEAAADQAVF GAFVQAGQSCVSTQRIFVHEDVVDAFTERMAAATRGLVVGDPLDETVDVSAVINEGEAIRVEKWIAEAEA GGAKVVVGGGRSGAVVTPTIIRGVDPAAKVQCQEVFGPVVTVNSFAEFTEALAAANDSVFGLNAGVFTTN LRHALQAIEELEFGSVYINDVPTVRADQQPYGGVKESGNTREGPRYAMEEMTELKFITLR
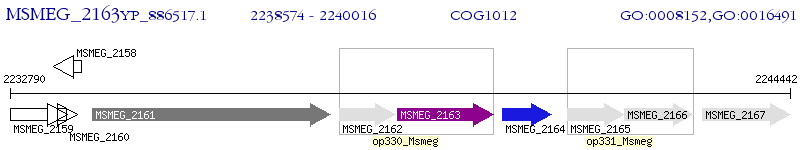
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2163 | - | - | 100% (480) | aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6297 | - | 3e-74 | 35.15% (478) | aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1665 | - | 1e-73 | 38.48% (473) | gamma-aminobutyraldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2883c | aldC | 6e-59 | 34.07% (452) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4860 | - | 6e-72 | 38.16% (477) | gamma-aminobutyraldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0223c | - | 3e-66 | 33.68% (478) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 2e-53 | 32.09% (455) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 5e-73 | 37.13% (474) | gamma-aminobutyraldehyde dehydrogenase |
| M. marinum M | MMAR_3556 | - | 2e-73 | 36.46% (480) | aldehyde dehydrogenase NAD dependent |
| M. avium 104 | MAV_1897 | - | 8e-72 | 36.36% (473) | betaine-aldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_4137 | - | 5e-72 | 35.98% (478) | PUTATIVE putative [NADP+] succinate-semialdehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_2789 | - | 2e-72 | 36.04% (480) | gamma-aminobutyraldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0014 | - | 5e-79 | 38.75% (480) | betaine-aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2883c|M.bovis_AF2122/97 ------------------------------MSTTQLINPATEEVLASVDH
Mvan_0014|M.vanbaalenii_PYR-1 ----------MSDIPHYRMYIDGAWRD--AADTIEVRSPATGELVATVAY
Rv0223c|M.tuberculosis_H37Rv --------MSDSATEYDKLFIGGKWTKPSTSDVIEVRCPATGEYVGKVPM
Mflv_4860|M.gilvum_PYR-GCK -------------MTVASSWIDGAPVK-TGGAQHTVINPATGESVAEFAL
MAB_3328|M.abscessus_ATCC_1997 -------------MSVPSSWINGRPVA-THGDTHRVINPATGEAVADLKL
MMAR_3556|M.marinum_M ---------MTTTVSVAGSWINGAAHS-TGGRQHAVINPATQETIAELAL
MUL_2789|M.ulcerans_Agy99 ---------MTTTVSVAGSWINGAAHS-TGGRQHAVINPATQETIAELAL
MAV_1897|M.avium_104 MTDTTISNGVTEQLVHFDLVIGGESVAAESGRTYDSIDPYTALPWARVPD
MSMEG_2163|M.smegmatis_MC2_155 -----MKGTQTMHDGPTAMYINGEWVAEGENGTTTLTAPFDGRELAVIPV
TH_4137|M.thermoresistible__bu ------------------MYIDGEWQSAASGKTFASINPANGAVLDSVPD
MLBr_02573|M.leprae_Br4923 -------------------------------MSIATINPATGETVKTFTP
* .
Mb2883c|M.bovis_AF2122/97 TDANAVDDAVQRARAAQRR--WARLAPAQRAAGLRAFAAAVQAHLDELAA
Mvan_0014|M.vanbaalenii_PYR-1 GGVTTVDDAVAAAKAADEAGVWRNTSAQQRADVLDAIADNLAARTDELTA
Rv0223c|M.tuberculosis_H37Rv AAAADVDAAVAAARAAFDNGPWPSTPPHERAAVIAAAVKMLAERKDLFTK
Mflv_4860|M.gilvum_PYR-GCK AQPGDVDRAVVSARAALPG--WKGATPAERSAVLAKLAKLADDATADLVA
MAB_3328|M.abscessus_ATCC_1997 ATTADVDAAVAAARAAGPA--WASSTPVDRSSVLAKLAAVLSANADTLVA
MMAR_3556|M.marinum_M ATPADVDRAVVAARRALPQ--WAGVTPAERSAVLAKLADLLEQRAPELIA
MUL_2789|M.ulcerans_Agy99 ATPADVDHAVVAARRALPQ--WAGVTTAERSAVLAELADLLEQRAPELIA
MAV_1897|M.avium_104 CGPADVDRAVAAARAALRG-PWGQLTGTARGKLLWRLGELIAREAESLAE
MSMEG_2163|M.smegmatis_MC2_155 GGRRDVERAVQAAHNARRT----GLPLDRRLTILEAVAASIAARSEDFAR
TH_4137|M.thermoresistible__bu GEREDARRAIDAAAAALPD--WRSRTAYERAEVLVEAHRIMLERRDDLAE
MLBr_02573|M.leprae_Br4923 TTQDEVEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEANQSAA
. *: * . *
Mb2883c|M.bovis_AF2122/97 LEVANSGHPIVSA-EWEAGHVRDVLAFYAASPE----RLSGRQIPVAG-G
Mvan_0014|M.vanbaalenii_PYR-1 LQVAENGATVRAAGAFLIGYAIAHLRYFAGLARTYAFQSSGPLMEAPTLA
Rv0223c|M.tuberculosis_H37Rv LLAAETGQPPTIIETMHWMGSMGAMNYFAGAAD----KVTWTETRTGSYG
Mflv_4860|M.gilvum_PYR-GCK EEVSQTGKPVRLATEFDVPGSVDNIDFFAGAAR---HLEGKASAEYSGDH
MAB_3328|M.abscessus_ATCC_1997 DEVAQTGKPVRLASEFDVPGSIDNVEFFAGAAR---HLEGKATAEYSADH
MMAR_3556|M.marinum_M EEVCQTGKPLRLAEEFDVPGSVDNIRFFAGAAR---RLPGQATAEYSADH
MUL_2789|M.ulcerans_Agy99 EEVCQTGKPLRLAEEFDVPGSVDNIRFFAGAAR---RLPGQATAEYSADH
MAV_1897|M.avium_104 LEVRDGGK-LVREMIGQMKALPDYYIYYAGLAD---KLQGEVVPVDKPNF
MSMEG_2163|M.smegmatis_MC2_155 AIALECAKPIKTARAEVLRAIDTIKFSVAAARTLGSESVPLDATAAGAGK
TH_4137|M.thermoresistible__bu LMTREQGKPLRMARTEVG-YAADFLLWFAEEAK---RVYGEVIPSARADQ
MLBr_02573|M.leprae_Br4923 LMTLEMGKTLQSAKDEAMKCAKGFRYYAEKAET--LLADEPAEAAAVGAS
. : .
Mb2883c|M.bovis_AF2122/97 VDVTFNEPMGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAELTPLT
Mvan_0014|M.vanbaalenii_PYR-1 AGMIVRDPVGVCAGMIPWNFPLLLAVWKLGPALAAGNTVVLKPDDQTPLT
Rv0223c|M.tuberculosis_H37Rv QSIVSREPVGVVGAIVAWNVPLFLAVNKIAPALLAGCTIVLKPAAETPLT
Mflv_4860|M.gilvum_PYR-GCK TSSIRREAVGVVATITPWNYPLQMAVWKVIPALAAGCSVVIKPAEITPLT
MAB_3328|M.abscessus_ATCC_1997 TSSIRREAAGVVATITPWNYPLQMAVWKVLPALAAGCTVVIKPSELTPLT
MMAR_3556|M.marinum_M TSSIRREAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTPLT
MUL_2789|M.ulcerans_Agy99 TSSIRREAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTPLT
MAV_1897|M.avium_104 FVYTRHEPVGVVGAITPWNSPLLLLTWKLAAGLAAGCTFVVKPSDHTPAS
MSMEG_2163|M.smegmatis_MC2_155 VTFTKRVPVGVVAAIAPFNFPLNLVVHKVAPALAVGCPVVLKPATQTPLS
TH_4137|M.thermoresistible__bu RFIVLAQPVGVCAAITPWNYPISMLTRKLAPALAAGCTSVLKPAEQTPLC
MLBr_02573|M.leprae_Br4923 KALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQC
. ** : .:* *: ..* .* ::* .*
Mb2883c|M.bovis_AF2122/97 TMRLGELAVEAGLDEDLLQVLPGKGTV-VGERFVTHPDIRKIVFTGSTEV
Mvan_0014|M.vanbaalenii_PYR-1 LLELARAADEVGLPAGVLNVVTGAGPT-VGARLAEHPDVRKVAFTGSTEV
Rv0223c|M.tuberculosis_H37Rv ANALAEVFAEVGLPEGVLSVVPG-GIE-TGQALTSNPDIDMFTFTGSSAV
Mflv_4860|M.gilvum_PYR-GCK TLTLARLASEAGLPDGVLNVVTGSGAD-VGAALAGHADVDMVTFTGSTPV
MAB_3328|M.abscessus_ATCC_1997 TLTLARLAAEAGLPEGVLNVVTGRGDD-VGYALASHPDVDLVTFTGSTAV
MMAR_3556|M.marinum_M TLTLARLAAEAGLPDGVLNVVTGLGAD-VGAALAAHPGVDLVTFTGSTAV
MUL_2789|M.ulcerans_Agy99 TLTLARLAAEAGLPDGVLNVVTGLGAD-VGAALAAHPGVDLVTFTGSTAV
MAV_1897|M.avium_104 TLAFAKLFAEAGFPPGVINVVTGWGPQ-TGAALAAHPDVDKIAFTGSTAT
MSMEG_2163|M.smegmatis_MC2_155 ALLLAEVFHEAGLPAGWLNVVCGSGAE-VGGALATHPDVAYVTFTGSPQV
TH_4137|M.thermoresistible__bu AVETFKVFADAGVPAGVVNLVTCSDPEPVADELLGDPRVRKISFTGSTEV
MLBr_02573|M.leprae_Br4923 ALYLADVIARGGFPEDCFQTLLISANG--VEAILRDPRVAAATLTGSEPA
*. . .. : : .. : :*** .
Mb2883c|M.bovis_AF2122/97 GKRVMAGAAAQVKRVTLELGGKSANIVFHDCDLERAATTAPAGVFDNAGQ
Mvan_0014|M.vanbaalenii_PYR-1 GKSVMRAAADTVKKVTLELGGKGASIVLDDADLDLAVDGSLFAFLLMSGQ
Rv0223c|M.tuberculosis_H37Rv GREVGRRAAEMLKPCTLELGGKSAAIILEDVDLAAAIPMMVFSGVMNAGQ
Mflv_4860|M.gilvum_PYR-GCK GRRVMLAAAAHGHRTQLELGGKAPFVVFDDADLDAAIQGAVAGALINTGQ
MAB_3328|M.abscessus_ATCC_1997 GRKVMAAAAVHGHRTQLELGGKAPFVVFDDADMDAAIAGAVAGAIINSGQ
MMAR_3556|M.marinum_M GRKVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGALINSGQ
MUL_2789|M.ulcerans_Agy99 GRKVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGAVINSGQ
MAV_1897|M.avium_104 GISVGKAAIDNMTRFTLELGGKSAQVVFDDADLDAAANGVIAGVFAATGQ
MSMEG_2163|M.smegmatis_MC2_155 GWGIAEAAPK--KKVRLELGSNAPLIVDRESDWEAAADQAVFGAFVQAGQ
TH_4137|M.thermoresistible__bu GRMIASRAGATMKRVSMELGGHAPFLVFPDADPEHAAKGGAAVKFLNTGQ
MLBr_02573|M.leprae_Br4923 GQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRVQNNGQ
* : * :***. . :: . * * . **
Mb2883c|M.bovis_AF2122/97 DCCARSRILVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMGPLVSRAHR
Mvan_0014|M.vanbaalenii_PYR-1 ACESGTRLLVHESIHDEFVRRMVARAETLVMGDPMSLASDLGPLVSAKQK
Rv0223c|M.tuberculosis_H37Rv GCVNQTRILAPRSRYDEIVAAVTNFVTALPVGPPSDPAAQIGPLISEKQR
Mflv_4860|M.gilvum_PYR-GCK DCTAATRAIVAEGLYDDFVAGVAEVMSKVVIGDPADPDTDLGPLISAVHR
MAB_3328|M.abscessus_ATCC_1997 DCTAATRALVARDLYDDFVAGVGEVMSKVVVGDPLDPDTDIGPLISAAHR
MMAR_3556|M.marinum_M DCTAATRAIVARELYHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLAHR
MUL_2789|M.ulcerans_Agy99 DCTAATRAIVARELCHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLAHR
MAV_1897|M.avium_104 TCLAGSRLLVHESVADALVERIVARASTIKLGDPKDPATEMGPVSNQPQY
MSMEG_2163|M.smegmatis_MC2_155 SCVSTQRIFVHEDVVDAFTERMAAATRGLVVGDPLDETVDVSAVINEGEA
TH_4137|M.thermoresistible__bu ACICPNRFIVHRSIAEPFIATLRARIEKLVPGDGLAAGTTVGPLIDEVAL
MLBr_02573|M.leprae_Br4923 SCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATESGR
* * :. . : . : * :..:
Mb2883c|M.bovis_AF2122/97 DKVAGYVP----DDAPVAFRGTAPAG----RGFWFPPTVLTPKRGD-RTV
Mvan_0014|M.vanbaalenii_PYR-1 ARVEKYIALGQEEGCKLAYQGTVPTDPALAQGHWVPPTILTGATNDMRIA
Rv0223c|M.tuberculosis_H37Rv TRVEGYIAKGIEEGARLVCGGGRPEG--LDNGFFIQPTVFADVDNKMTIA
Mflv_4860|M.gilvum_PYR-GCK EKVAGMVERAPGEGGRVVTGGVAPDG----PGSFYRPTLIADVAESSEVW
MAB_3328|M.abscessus_ATCC_1997 AKVSSIVDRAPAQGGRIVTGATAPDL----PGSFYRPTLIADVAETSEVY
MMAR_3556|M.marinum_M DRVADIVAKAPAQGARIVVGGRIPDL----AGAFYLPTLLADVGEQSQAY
MUL_2789|M.ulcerans_Agy99 DRVADILAKAPAQGARIVVGGRIPDL----AGAFYLPTLLADVGEQSQAY
MAV_1897|M.avium_104 EKVLSHFASAREQGATVAYGGE-PAG--ELGGFFVKPTVLTGVDRSMRAV
MSMEG_2163|M.smegmatis_MC2_155 IRVEKWIAEAEAGGAKVVVGGGRSGA-------VVTPTIIRGVDPAAKVQ
TH_4137|M.thermoresistible__bu KKMQRQVDDAVDKGAKLELGGSRLQGPAFDNGHFYAPTILTGVTPDMLIC
MLBr_02573|M.leprae_Br4923 AQVEQQVDAAAAAGAVIRCGGKRPDR----PGWFYPPTVITDITKDMALY
:: . . : . **::
Mb2883c|M.bovis_AF2122/97 TDEIFGPVVVVLTFDDEADAISLANDTAYGLSGSIWTDDLSRALRVARAV
Mvan_0014|M.vanbaalenii_PYR-1 REEIFGPVLVVLTYGDDDEAVAIANDSEYGLSAGVWSADRERALGIARRL
Rv0223c|M.tuberculosis_H37Rv QEEIFGPVLAIIPYDTEEDAIAIANDSVYGLAGSVWTTDVPKGIKISQQI
Mflv_4860|M.gilvum_PYR-GCK RDEIFGPVLTVRSFTDDDDAIRQANDTKYGLAASAWTRDVYRAQRASREI
MAB_3328|M.abscessus_ATCC_1997 RDEIFGPVLTVRPFTDDDDALRQANDTAYGLAASAWTRDVYRAQRASREI
MMAR_3556|M.marinum_M REEIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREI
MUL_2789|M.ulcerans_Agy99 REEIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREI
MAV_1897|M.avium_104 AEEVFGPVLAVMTFTDEDEAIAAANDTEFGLAAGVWTKDVHRAHRVAARL
MSMEG_2163|M.smegmatis_MC2_155 CQEVFGPVVTVNSFAEFTEALAAANDSVFGLNAGVFTTNLRHALQAIEEL
TH_4137|M.thermoresistible__bu SEETFGPIAPVMVFDDEDEAIAMANSTEYGLAGYIYTRDISRAIRVGEAL
MLBr_02573|M.leprae_Br4923 TDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFIDNI
:* ***: : :*: ** : :** . :: : . :
Mb2883c|M.bovis_AF2122/97 ESGNLSVNSHSSVRF-NTPFGGFKQSGVGRELGPDAPLQFTETKNVFIAV
Mvan_0014|M.vanbaalenii_PYR-1 QSGTVWVNDWHMINA-MYPFGGVKQSGLGRELGPDALDEYTEPKFIHVDM
Rv0223c|M.tuberculosis_H37Rv RTGTYGIN-WYAFDP-GSPFGGYKNSGIGRENGPEGVEHFTQQKSVLLPM
Mflv_4860|M.gilvum_PYR-GCK HAGCVWINDHIPIIS-EMPHGGFGASGFGKDMSQYSLEEYLSIKHVMSDI
MAB_3328|M.abscessus_ATCC_1997 RAGCVWINDHIPIIS-EMPHGGFGASGFGKDMSDYSLEEYLTVKHVMSDI
MMAR_3556|M.marinum_M SAGCVWINDHIPIVS-EMPHGGVAASGFGKDMSEYSFEEYLTIKHVMSDI
MUL_2789|M.ulcerans_Agy99 SAGCVWINDHIPIVS-EMPHGGVAASGFGNDMSEYSFDEYLTIKHVMSDI
MAV_1897|M.avium_104 NAGTVWVNAYRVVSP-HVPFGGMGHSGIGRENGIDAVKDFTETKAVWVEL
MSMEG_2163|M.smegmatis_MC2_155 EFGSVYINDVPTVRADQQPYGGVKESGNTREGPRYAMEEMTELKFITLR-
TH_4137|M.thermoresistible__bu DFGMIGINDINPTSA-AVPFGGIKQSGLGREGARAGIEEYLDTKVLGIAL
MLBr_02573|M.leprae_Br4923 VAGQVFINGMTVSYP-ELPFGGIKRSGYGRELSTHGIREFCNIKTVWIA-
* :* *.** ** .: . . * :
Mb2883c|M.bovis_AF2122/97 GEEM---------------
Mvan_0014|M.vanbaalenii_PYR-1 TDDRRKHVYPVVISAAAQG
Rv0223c|M.tuberculosis_H37Rv GYTVA--------------
Mflv_4860|M.gilvum_PYR-GCK TSAVDKDWHRTIFTKR---
MAB_3328|M.abscessus_ATCC_1997 TGTADKDWHRTIFAKR---
MMAR_3556|M.marinum_M TGVAEKPWHRTIFSTRI--
MUL_2789|M.ulcerans_Agy99 TGVAEKPWHRTIFSTRI--
MAV_1897|M.avium_104 SGATRDPFTLG--------
MSMEG_2163|M.smegmatis_MC2_155 -------------------
TH_4137|M.thermoresistible__bu -------------------
MLBr_02573|M.leprae_Br4923 -------------------